Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo - PubMed (original) (raw)
Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo
Huabin Zhao et al. Mol Biol Evol. 2010 Dec.
Abstract
Although it belongs to the order Carnivora, the giant panda is a vegetarian with 99% of its diet being bamboo. The draft genome sequence of the giant panda shows that its umami taste receptor gene Tas1r1 is a pseudogene, prompting the proposal that the loss of the umami perception explains why the giant panda is herbivorous. To test this hypothesis, we sequenced all six exons of Tas1r1 in another individual of the giant panda and five other carnivores. We found that the open reading frame (ORF) of Tas1r1 is intact in all these carnivores except the giant panda. The rate ratio (ω) of nonsynonymous to synonymous substitutions in Tas1r1 is significantly higher for the giant panda lineage than for other carnivore lineages. Based on the ω change and the observed number of ORF-disrupting substitutions, we estimated that the functional constraint on the giant panda Tas1r1 was relaxed ∼ 4.2 Ma, with its 95% confidence interval between 1.3 and 10 Ma. Our estimate matches the approximate date of the giant panda's dietary switch inferred from fossil records. It is probable that the giant panda's decreased reliance on meat resulted in the dispensability of the umami taste, leading to Tas1r1 pseudogenization, which in turn reinforced its herbivorous life style because of the diminished attraction of returning to meat eating in the absence of Tas1r1. Nonetheless, additional factors are likely involved because herbivores such as cow and horse still retain an intact Tas1r1.
Figures
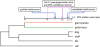
FIG. 1.
A species tree of the carnivores studied in this work. Branch lengths are not drawn to scale except for the giant panda branch. Dates indicated by blue arrows are inferred from fossil records, whereas dates indicated with the purple arrow and dash lines are inferred from the molecular genetic evidence of this study.
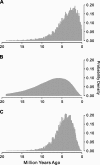
FIG. 2.
Posterior probability distributions of the time at which the functional constraint on the giant panda Tas1r1 was completely relaxed, based on the observations from (A) the rate of nucleotide substitutions, (B) the number of ORF-disrupting substitutions, and (C) both.
Similar articles
- Why does the giant panda eat bamboo? A comparative analysis of appetite-reward-related genes among mammals.
Jin K, Xue C, Wu X, Qian J, Zhu Y, Yang Z, Yonezawa T, Crabbe MJ, Cao Y, Hasegawa M, Zhong Y, Zheng Y. Jin K, et al. PLoS One. 2011;6(7):e22602. doi: 10.1371/journal.pone.0022602. Epub 2011 Jul 27. PLoS One. 2011. PMID: 21818345 Free PMC article. - Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas.
Hu Y, Wu Q, Ma S, Ma T, Shan L, Wang X, Nie Y, Ning Z, Yan L, Xiu Y, Wei F. Hu Y, et al. Proc Natl Acad Sci U S A. 2017 Jan 31;114(5):1081-1086. doi: 10.1073/pnas.1613870114. Epub 2017 Jan 17. Proc Natl Acad Sci U S A. 2017. PMID: 28096377 Free PMC article. - Comparative Analysis of the Umami Taste Receptor Gene Tas1r1 in Mustelidae.
Tarusawa Y, Matsumura S. Tarusawa Y, et al. Zoolog Sci. 2020 Apr;37(2):122-127. doi: 10.2108/zs190086. Zoolog Sci. 2020. PMID: 32282143 - Lineage-specific evolution of bitter taste receptor genes in the giant and red pandas implies dietary adaptation.
Shan L, Wu Q, Wang L, Zhang L, Wei F. Shan L, et al. Integr Zool. 2018 Mar;13(2):152-159. doi: 10.1111/1749-4877.12291. Integr Zool. 2018. PMID: 29168616 Free PMC article. - Dietary resources shape the adaptive changes of cyanide detoxification function in giant panda (Ailuropoda melanoleuca).
Huang H, Yie S, Liu Y, Wang C, Cai Z, Zhang W, Lan J, Huang X, Luo L, Cai K, Hou R, Zhang Z. Huang H, et al. Sci Rep. 2016 Oct 5;6:34700. doi: 10.1038/srep34700. Sci Rep. 2016. PMID: 27703267 Free PMC article.
Cited by
- L-Glutamate Regulates Npy via the mGluR4-Ca2+-ERK1/2 Signaling Pathway in Mandarin Fish (Siniperca chuatsi).
Duan J, Wang Q, He S, Liang XF, Ding L. Duan J, et al. Int J Mol Sci. 2024 Sep 18;25(18):10035. doi: 10.3390/ijms251810035. Int J Mol Sci. 2024. PMID: 39337521 Free PMC article. - Pseudogenes in plasmid genomes reveal past transitions in plasmid mobility.
Hanke DM, Wang Y, Dagan T. Hanke DM, et al. Nucleic Acids Res. 2024 Jul 8;52(12):7049-7062. doi: 10.1093/nar/gkae430. Nucleic Acids Res. 2024. PMID: 38808675 Free PMC article. - A chromosome-level genome of electric catfish (Malapterurus electricus) provided new insights into order Siluriformes evolution.
Liu M, Song Y, Zhang S, Yu L, Yuan Z, Yang H, Zhang M, Zhou Z, Seim I, Liu S, Fan G, Yang H. Liu M, et al. Mar Life Sci Technol. 2023 Dec 14;6(1):1-14. doi: 10.1007/s42995-023-00197-8. eCollection 2024 Feb. Mar Life Sci Technol. 2023. PMID: 38433969 Free PMC article. - Diversity and evolution of the vertebrate chemoreceptor gene repertoire.
Policarpo M, Baldwin MW, Casane D, Salzburger W. Policarpo M, et al. Nat Commun. 2024 Feb 15;15(1):1421. doi: 10.1038/s41467-024-45500-y. Nat Commun. 2024. PMID: 38360851 Free PMC article.
References
- Arnason U, Gullberg A, Janke A, Kullberg M. Mitogenomic analyses of caniform relationships. Mol Phylogenet Evol. 2007;45:863–874. - PubMed
- Bininda-Emonds OR. Phylogenetic position of the giant panda. In: Lindburg D, Baragona K, editors. Giant panda: biology and conservation. Berkeley (CA): University of California Press; 2004.
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. - PubMed
- Goldman DP, Giri R, J O'Brien S. Molecular genetic-distance estimates among the Ursidae as indicated by one- and two-dimensional protein electrophoresis. Evolution. 1989;43:282–295. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources