Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells - PubMed (original) (raw)
Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells
Marion Dejosez et al. Genes Dev. 2010.
Abstract
Self-renewing embryonic stem (ES) cells have an exceptional need for timely biomass production, yet the transcriptional control mechanisms responsible for meeting this requirement are largely unknown. We report here that Ronin (Thap11), which is essential for the self-renewal of ES cells, binds with its transcriptional coregulator, Hcf-1, to a highly conserved enhancer element that previously lacked a recognized binding factor. The subset of genes bound by Ronin/Hcf-1 function primarily in transcription initiation, mRNA splicing, and cell metabolism; genes involved in cell signaling and cell development are conspicuously underrepresented in this target gene repertoire. Although Ronin/Hcf-1 represses the expression of some target genes, its activity at promoter sites more often leads to the up-regulation of genes essential to protein biosynthesis and energy production. We propose that Ronin/Hcf-1 controls a genetic program that contributes to the unimpeded growth of ES cells.
Figures
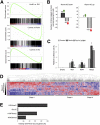
Figure 1.
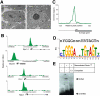
Ronin binds to a hyperconserved enhancer element in mouse ES cells. (A) _Ronin_-overexpressing clones are characterized by a rounder cell shape and a more prominent nucleolus than found in control clones (electronmicroscopy; bar, 2 μm). (B) Binding of Ronin at the promoter regions of four representative genes. ChIP-seq results, shown on the _Y_-axis, are reads per million reads. (C) Histogram showing the distance of the midpoint of each Ronin-binding event from the nearest TSS (arrow). (D) Identification of the consensus RBM depicted as a bit matrix. (Top) The recently discovered M4 sequence in human promoters (Xie et al. 2005) is included for comparison. (E) EMSA analysis of the newly identified RBM using recombinant Ronin1–90 (Thap domain). The specific shift of the RBM is abolished in the presence of a specific competitor (C1).
Figure 2.
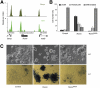
Ronin and Hcf-1 bind together to specific genes. (A) ChIP-seq results obtained in mouse ES cells after immunoprecipitation with Ronin or Hcf-1 antibodies. A representative region containing three Ronin-bound genes shows substantial overlap between Ronin- and Hcf-1-binding peaks in the promoter regions of all three genes. (B) Quantification of the experiment shown in the bottom panel of C. Values are means ± SD of triplicate experiments. (C, top panel) Morphology of control, _Ronin_-overexpressing, and _RoninDHSA_-overexpressing mouse ES cells after 3 d of culture in the presence of Lif (10× magnification). (Bottom panel) Cells were stained for alkaline phosphatase activity after 4 d of culture in the absence of Lif (20× magnification).
Figure 3.
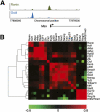
Comparison of binding characteristics of Ronin and canonical pluripotency factors. (A) ChIP-seq results obtained by precipitation with antibodies against Ronin or Oct4. (B) Hierarchical clustering analysis of 22 prominent transcriptional regulators in mouse ES cells, based on target similarity scores calculated with a Pearson correlation similarity metric.
Figure 4.
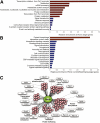
Cellular functions of genes targeted by Ronin/Hcf-1 in mouse ES cells. (A) PANTHER analysis of target genes. Categories with enrichment values >1 are significantly overrepresented (red), while those with lower values are significantly underrepresented (blue). (B) PANTHER analysis of Ronin versus Oct4/Sox2/Nanog targets, as in A. (C) Summary of Ronin/Hcf-1 target genes by major functional categories.
Figure 5.
Ronin/Hcf-1 can either activate or repress its transcriptional targets in mouse ES cells. (A) GSEA analyses of Ronin/Hcf-1 target genes, showing enrichment of bound genes in undifferentiated versus differentiated mouse ES cells (top panel), in _Ronin_-overexpressing versus control ES cells (middle panel), and in control compared with Ronin knockout ES cells (bottom panel). (B) Overlap between the numbers of Ronin-bound genes that are up-regulated or down-regulated in _Ronin_-overexpressing ES cells in relation to their status in Ronin knockout ES cells. (C) Luciferase reporter assays with pGL3-based promoter constructs, showing effects on the expression of selected target genes. (*) P < 0.05; (**) P < 0.01. (ΔRBM) Mutant reporter construct with deleted RBM. (D) Relative expression levels of Ronin target genes during the differentiation of mouse ES cells. (Red) Up-regulated genes; (blue) down-regulated genes. (E) Prevalence of H3K4me3-, Suz12-, H3K79me2-, or H3K36me3-enriched regions at Ronin-binding sites.
Similar articles
- Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells.
Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Dejosez M, et al. Cell. 2008 Jun 27;133(7):1162-74. doi: 10.1016/j.cell.2008.05.047. Cell. 2008. PMID: 18585351 Free PMC article. - Ronin Governs Early Heart Development by Controlling Core Gene Expression Programs.
Fujita J, Freire P, Coarfa C, Benham AL, Gunaratne P, Schneider MD, Dejosez M, Zwaka TP. Fujita J, et al. Cell Rep. 2017 Nov 7;21(6):1562-1573. doi: 10.1016/j.celrep.2017.10.036. Cell Rep. 2017. PMID: 29117561 Free PMC article. - Ronin influences the DNA damage response in pluripotent stem cells.
Seifert BA, Dejosez M, Zwaka TP. Seifert BA, et al. Stem Cell Res. 2017 Aug;23:98-104. doi: 10.1016/j.scr.2017.06.014. Epub 2017 Jul 3. Stem Cell Res. 2017. PMID: 28715716 Free PMC article. - Role of host cell factor-1 in cell cycle regulation.
Zargar Z, Tyagi S. Zargar Z, et al. Transcription. 2012 Jul-Aug;3(4):187-92. doi: 10.4161/trns.20711. Epub 2012 Jul 1. Transcription. 2012. PMID: 22771988 Free PMC article. Review. - Nanog and transcriptional networks in embryonic stem cell pluripotency.
Pan G, Thomson JA. Pan G, et al. Cell Res. 2007 Jan;17(1):42-9. doi: 10.1038/sj.cr.7310125. Cell Res. 2007. PMID: 17211451 Review.
Cited by
- Embryonic stem cell interactomics: the beginning of a long road to biological function.
Yousefi M, Hajihoseini V, Jung W, Hosseinpour B, Rassouli H, Lee B, Baharvand H, Lee K, Salekdeh GH. Yousefi M, et al. Stem Cell Rev Rep. 2012 Dec;8(4):1138-54. doi: 10.1007/s12015-012-9400-9. Stem Cell Rev Rep. 2012. PMID: 22847281 Review. - O-GlcNAc cycling: a link between metabolism and chronic disease.
Bond MR, Hanover JA. Bond MR, et al. Annu Rev Nutr. 2013;33:205-29. doi: 10.1146/annurev-nutr-071812-161240. Epub 2013 Apr 29. Annu Rev Nutr. 2013. PMID: 23642195 Free PMC article. Review. - THAP and ATF-2 regulated sterol carrier protein-2 promoter activities in the larval midgut of the yellow fever mosquito, Aedes aegypti.
Peng R, Fu Q, Hong H, Schwaegler T, Lan Q. Peng R, et al. PLoS One. 2012;7(10):e46948. doi: 10.1371/journal.pone.0046948. Epub 2012 Oct 4. PLoS One. 2012. PMID: 23056538 Free PMC article. - Hcfc1b, a zebrafish ortholog of HCFC1, regulates craniofacial development by modulating mmachc expression.
Quintana AM, Geiger EA, Achilly N, Rosenblatt DS, Maclean KN, Stabler SP, Artinger KB, Appel B, Shaikh TH. Quintana AM, et al. Dev Biol. 2014 Dec 1;396(1):94-106. doi: 10.1016/j.ydbio.2014.09.026. Epub 2014 Oct 2. Dev Biol. 2014. PMID: 25281006 Free PMC article. - Abnormal chondrocyte development in a zebrafish model of cblC syndrome restored by an MMACHC cobalamin binding mutant.
Paz D, Pinales BE, Castellanos BS, Perez I, Gil CB, Madrigal LJ, Reyes-Nava NG, Castro VL, Sloan JL, Quintana AM. Paz D, et al. Differentiation. 2023 May-Jun;131:74-81. doi: 10.1016/j.diff.2023.04.003. Epub 2023 May 5. Differentiation. 2023. PMID: 37167860 Free PMC article.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 - PubMed
- Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, Lazar V, Dessen P, Mantovani R, Aguilar L, et al. 2007. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109: 584–594 - PubMed
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2R01 HG002668/HG/NHGRI NIH HHS/United States
- P01 GM081627/GM/NIGMS NIH HHS/United States
- R01 HG002668/HG/NHGRI NIH HHS/United States
- P20 EB007076/EB/NIBIB NIH HHS/United States
- 1R01 GM077442-01/GM/NIGMS NIH HHS/United States
- R01 EB005173/EB/NIBIB NIH HHS/United States
- R01 GM077442/GM/NIGMS NIH HHS/United States
- P01 GM81627/GM/NIGMS NIH HHS/United States
- R01 EB005173-01/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials