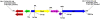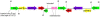Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences - PubMed (original) (raw)
Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences
Annemieke Smet et al. PLoS One. 2010.
Abstract
Background: CTX-M-producing Escherichia coli strains are regarded as major global pathogens.
Methodology/principal findings: The nucleotide sequence of three plasmids (pEC_B24: 73801-bp; pEC_L8: 118525-bp and pEC_L46: 144871-bp) from Escherichia coli isolates obtained from patients with urinary tract infections and one plasmid (pEC_Bactec: 92970-bp) from an Escherichia coli strain isolated from the joint of a horse with arthritis were determined. Plasmid pEC_Bactec belongs to the IncI1 group and carries two resistance genes: bla(TEM-1) and bla(CTX-M-15). It shares more than 90% homology with a previously published bla(CTX-M)-plasmid from E. coli of human origin. Plasmid pEC_B24 belongs to the IncFII group whereas plasmids pEC_L8 and pEC_L46 represent a fusion of two replicons of type FII and FIA. On the pEC_B24 backbone, two resistance genes, bla(TEM-1) and bla(CTX-M-15), were found. Six resistance genes, bla(TEM-1), bla(CTX-M-15), bla(OXA-1), aac6'-lb-cr, tetA and catB4, were detected on the pEC_L8 backbone. The same antimicrobial drug resistance genes, with the exception of tetA, were also identified on the pEC_L46 backbone. Genome analysis of all 4 plasmids studied provides evidence of a seemingly frequent transposition event of the bla(CTX-M-15)-ISEcp1 element. This element seems to have a preferred insertion site at the tnpA gene of a bla(TEM)-carrying Tn3-like transposon, the latter itself being inserted by a transposition event. The IS26-composite transposon, which contains the bla(OXA-1), aac6'-lb-cr and catB4 genes, was inserted into plasmids pEC_L8 and pEC_L46 by homologous recombination rather than a transposition event. Results obtained for pEC_L46 indicated that IS26 also plays an important role in structural rearrangements of the plasmid backbone and seems to facilitate the mobilisation of fragments from other plasmids.
Conclusions: Collectively, these data suggests that IS26 together with ISEcp1 could play a critical role in the evolution of diverse multiresistant plasmids found in clinical Enterobacteriaceae.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
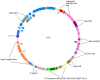
Figure 1. Physical map of pEC_Bactec (IncI1).
The hypothetical proteins and predicted ORFs are presented by coloured boxes (blue for the tra locus, orange for the type IV prepilin cluster and pink for hypothetical proteins). The I1 replicon is coloured in grey. The tnpA genes are indicated by red boxes whereas the IS_26_ element is bright green. Other genes that have an important function for pEC_Bactec bear different coloured boxes: green-brown, impC/A/B genes; blue-grey, sogL/sogS genes; blue-green, psiA/psiB genes; dark pink, ardA gene: purple, nikA/nikB genes; blue-purple, rci gene; red-brown, klcA gene; and dark-grey, pndA/pndC. The new Tn_3_ element, containing tnpA (green) and tnpR (grey) genes and encoding CTX-M-15 (green) in addition to TEM-1 (yellow) is also highlighted. The IS_Ecp1_ element linked to bla CTX-M-15 is coloured in orange.
Figure 2. Detail of the new Tn_3_ element, containing tnpA (disrupted, blue), tnpR (green), orf477 (purple), bla CTX-M-15 (pink), IS_Ecp1_ (yellow) and bla TEM-1 or bla TEM-33 (red) genes.
The 5-bp direct repeats consistent with transposition events of the Tn_3_ transposon and the IS_Ecp1_- bla CTX-M-15 element are also shown, as are tags for inverted repeats (IR prefix) and basepair numbering corresponding to transposon-seperated segments of the tnpA gene.
Figure 3. Physical maps of pEC_B24 (IncFII), pEC_L8 (IncFII, FIA) and pEC_L46 (IncFII, FIA).
The hypothetical proteins and predicted ORFs are represented by coloured boxes. The pEC_L8 (inner circle) is compared with pEC_L46 (outer circle). Dashed lines stake out a large common region (right half). The tra locus is indicated by blue boxes and the hypothetical proteins are indicated in pink. The tnpA genes are indicated in red boxes whereas the IS_26_ element is coloured in bright green. The antitoxin/toxin genes are indicated by green-brown (vagC/vagD), purple (pemI/pemK), blue-grey (ccdA/ccdB) and brown-black-red (hok/mok, parB, psiA/psiB), respectively. The FII replicon and FIA replicon is indicated in grey and grey-green, respectively. Other genes that have an important function for these IncF plasmids are indicated by different coloured boxes: dark blue (ugpB, ugpC, araQ and ugpA), dark green (icc, klcA), green (yigB), purple (sopB, kdgT), orange (yihH, tdcF), orange-pink (colicin B/M genes) and purple blue (yfaX). The new Tn_3_ element, containing tnpA (green) and tnpR (grey) genes and encoding CTX-M-15 (green) in addition to TEM-1 (yellow) is also highlighted. The IS_Ecp1_ element linked to bla CTX-M-15 is coloured orange. The IS_26_ (bright-green) composite transposon carrying aac6'-lb-cr (purple-blue), bla OXA-1 (light yellow) and catB4 (pink) is also shown as is the pEC_L46 fragment with more than 90% homology to a segment from a Klebsiella plasmid.
Figure 4. Detail of two IS_26_-composite transposon elements carrying the bla OXA-1, aac6'-lb-cr and catB4 genes, which are inserted in an inverted orientation.
Both elements are separated by two oxidoreductase (yigB genes) elements. The intact left and right inverted repeats of IS_26_ are also shown
Similar articles
- Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone.
Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. Woodford N, et al. Antimicrob Agents Chemother. 2009 Oct;53(10):4472-82. doi: 10.1128/AAC.00688-09. Epub 2009 Aug 17. Antimicrob Agents Chemother. 2009. PMID: 19687243 Free PMC article. - Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements.
Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. Zong Z, et al. Plasmid. 2015 Jul;80:118-26. doi: 10.1016/j.plasmid.2015.04.007. Epub 2015 Apr 27. Plasmid. 2015. PMID: 25929173 - A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates.
Cullik A, Pfeifer Y, Prager R, von Baum H, Witte W. Cullik A, et al. J Med Microbiol. 2010 May;59(Pt 5):580-587. doi: 10.1099/jmm.0.016188-0. Epub 2010 Jan 21. J Med Microbiol. 2010. PMID: 20093380 - Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada.
Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. Boyd DA, et al. Antimicrob Agents Chemother. 2004 Oct;48(10):3758-64. doi: 10.1128/AAC.48.10.3758-3764.2004. Antimicrob Agents Chemother. 2004. PMID: 15388431 Free PMC article. - ISEcp1-mediated transposition of linked blaCTX-M-3 and blaTEM-1b from the IncI1 plasmid pEK204 found in clinical isolates of Escherichia coli from Belfast, UK.
Dhanji H, Doumith M, Hope R, Livermore DM, Woodford N. Dhanji H, et al. J Antimicrob Chemother. 2011 Oct;66(10):2263-5. doi: 10.1093/jac/dkr310. Epub 2011 Jul 26. J Antimicrob Chemother. 2011. PMID: 21795257
Cited by
- Complete nucleotide sequences of bla(CTX-M)-harboring IncF plasmids from community-associated Escherichia coli strains in the United States.
Li JJ, Spychala CN, Hu F, Sheng JF, Doi Y. Li JJ, et al. Antimicrob Agents Chemother. 2015;59(6):3002-7. doi: 10.1128/AAC.04772-14. Epub 2015 Mar 9. Antimicrob Agents Chemother. 2015. PMID: 25753630 Free PMC article. - Epidemic plasmid carrying bla(CTX-M-15) in Klebsiella penumoniae in China.
Zhuo C, Li XQ, Zong ZY, Zhong NS. Zhuo C, et al. PLoS One. 2013;8(1):e52222. doi: 10.1371/journal.pone.0052222. Epub 2013 Jan 29. PLoS One. 2013. PMID: 23382815 Free PMC article. - Epidemiology of Plasmid Lineages Mediating the Spread of Extended-Spectrum Beta-Lactamases among Clinical Escherichia coli.
Mahmud B, Wallace MA, Reske KA, Alvarado K, Muenks CE, Rasmussen DA, Burnham CD, Lanzas C, Dubberke ER, Dantas G. Mahmud B, et al. mSystems. 2022 Oct 26;7(5):e0051922. doi: 10.1128/msystems.00519-22. Epub 2022 Aug 22. mSystems. 2022. PMID: 35993734 Free PMC article. - Food Workers as a Reservoir of Extended-Spectrum-Cephalosporin-Resistant Salmonella Strains in Japan.
Shigemura H, Sakatsume E, Sekizuka T, Yokoyama H, Hamada K, Etoh Y, Carle Y, Mizumoto S, Hirai S, Matsui M, Kimura H, Suzuki M, Onozuka D, Kuroda M, Inoshima Y, Murakami K. Shigemura H, et al. Appl Environ Microbiol. 2020 Jun 17;86(13):e00072-20. doi: 10.1128/AEM.00072-20. Print 2020 Jun 17. Appl Environ Microbiol. 2020. PMID: 32276982 Free PMC article. - Initial Assessment of the Molecular Epidemiology of blaNDM-1 in Colombia.
Rojas LJ, Wright MS, De La Cadena E, Motoa G, Hujer KM, Villegas MV, Adams MD, Bonomo RA. Rojas LJ, et al. Antimicrob Agents Chemother. 2016 Jun 20;60(7):4346-50. doi: 10.1128/AAC.03072-15. Print 2016 Jul. Antimicrob Agents Chemother. 2016. PMID: 27067339 Free PMC article.
References
- Bywater RJ. Veterinary use of antimicrobials and emergence of resistance in zoonotic and sentinel bacteria in the EU. J Vet Med. 2004;51:361–363. - PubMed
- Bywater RJ. Identification and surveillance of antimicrobial resistance dissemination in animal production. Poultry Sci. 2005;84:644–648. - PubMed
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. - PubMed
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases