Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO - PubMed (original) (raw)
Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO
Taiyun Wei et al. PLoS Pathog. 2010.
Abstract
Intercellular transport of viruses through cytoplasmic connections, termed plasmodesmata (PD), is essential for systemic infection in plants by viruses. Previous genetic and ultrastructural data revealed that the potyvirus cyclindrical inclusion (CI) protein is directly involved in cell-to-cell movement, likely through the formation of conical structures anchored to and extended through PD. In this study, we demonstrate that plasmodesmatal localization of CI in N. benthamiana leaf cells is modulated by the recently discovered potyviral protein, P3N-PIPO, in a CI:P3N-PIPO ratio-dependent manner. We show that P3N-PIPO is a PD-located protein that physically interacts with CI in planta. The early secretory pathway, rather than the actomyosin motility system, is required for the delivery of P3N-PIPO and CI to PD. Moreover, CI mutations that disrupt virus cell-to-cell movement compromise PD-localization capacity. These data suggest that the CI and P3N-PIPO complex coordinates the formation of PD-associated structures that facilitate the intercellular movement of potyviruses in infected plants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
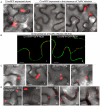
Figure 1. Schematic representation of the TuMV genome.
The circle represents the genome-linked viral protein, VPg. Two short horizontal lines represent 5′ and 3′ untranslational region, respectively. The large box represents the long open reading frame (from nucleotides 131 to 9625). The mature proteins resulting from processing the large polyprotein are indicated as smaller boxes. PIPO (from nucleotides 3079 to 3258) derived from a frameshif on the P3 cistron is indicated as a shot grey bar. P3N-PIPO is indicated in green and CI in red. The poly(A) tail is shown as (A)n. For clarity, the relative sizes of the mature proteins are not drawn to scale.
Figure 2. Subcellular localization of TuMV CI in N. benthamiana leaf cells.
(A) Localization of CI-mRFP expressed alone in the leaf cells 48 hrs post-agroinfiltration (panel I) or in TuMV-infected leaf tissues 48 hrs (panel II) or 72 hrs (panel III) post-agroinfiltration. (B) Localization of CI-mRFP coexpressing with the plasma membrane marker GFP-REM (panels I, II) 48 hrs post-agroinfiltration. (C) Localization of CI-mRFP in the leaf cells coexpressing other viral proteins, i.e., HC-Pro (panel I), P3 (panel II), VPg (panel III) or CP (panel IV). Images were taken 48 hrs post-agroinfiltration. (D) Localization of CI-mRFP in the cells coexpressing P3N-PIPO. The ratio of agrobacterial culture mixtures containing plasmid CI to plasmid P3N-PIPO is indicated. Bars, 8 µm.
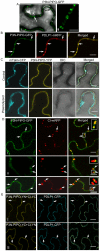
Figure 3. TuMV P3N-PIPO is a PD-localized protein and mediates the targeting of CI to PD in N. benthamiana.
(A, panels I, II) Localization of P3N-PIPO-GFP transiently expressed in the cell treated 48 hrs post-agroinfiltration. Paired P3N-PIPO structures under a higher magnification (panel II). (B) Colocalization of P3N-PIPO-GFP with the PD marker PDLP1-mRFP. Arrows point to PD costained by P3N-PIPO-GFP and PDLP1-mRFP. (C) Cells coexpressing P3N-PIPO-YFP and mTalin-CFP as a cell membrane marker (control). Fluorescence of P3N-PIPO-YFP in plasmolyzed leaf tissue containing mTalin-CFP (plasmolyzed). DIC, Differential interference contrast. (D) Colocalization of P3N-PIPO with CI-mRFP 48 hrs (panel I) and 72 hrs (panels II, III) post-agroinfiltration. Arrows point to the PD-localized P3N-PIPO-GFP and CI-mRFP. Insets are the enlarged images of the areas in white boxes in the corresponding panels. (E) Interactions of TuMV CI and P3N-PIPO proteins in vivo. BiFC analysis (48 hrs post-agroinfiltration) was used to assess interactions in cells coexpressing CI-YC and P3N-PIPO-YN (panel I), CI-YN and P3N-PIPO-YC (panel II). Arrows indicate the strong BiFC fluorescence at PD costained by the PD marker, PDLP1-CFP. Bars, 8 µm.
Figure 4. Targeting of P3N-PIPO and CI to PD requires the BFA-sensitive secretory pathway and is independent of the acto-myosin motility system.
PD marker PDLP-1-CFP (panels I), P3N-PIPO-YFP (panels II) and CI-mRFP in the presence of untagged P3N-PIPO (panels III) were transiently expressed in leaf cells treated with water (Control, A), 50 µg/mL BFA (BFA, B), co-agroinfiltrated with the untagged COPII mutant Sar1(H74L) [Sar1(H74L), C], co-agroinfiltrated with the untagged myosin XI-K tail (Myosin XI-K tail, D), co-agroinfiltrated with the untagged myosin VIII-1 tail (Myosin VIII-1 tail, E), and 25 µM Lat B (Lat B, F). Images were taken 48 hrs post-agroinfiltration. N, nucleus. Bars, 10 µm.
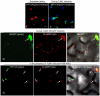
Figure 5. Association of TuMV CP with CI in TuMV-infected N. benthamiana leaf cells.
(Panels I, II) TuMV CP tagged by YFP (YFP-CP) in the cytoplasm when expressed alone (panel I) or during virus infection (panel II). (Panel III) When coexpressed with the recombinant TuMV::6K-GFP infectious clone, some mRFP-CP is also present in proximity to the 6K-GFP-labeled replication complex (arrow). (Panel IV) TuMV YFP-CP attachment to PD-associated CI structures.(arrows; Inset) in the cell periphery in the presence of P3N-PIPO during virus infection. All images are taken 48 hrs post-agroinfiltration. Chl, chloroplasts. Bars, 8 _µ_m.
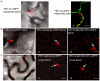
Figure 6. Subcellular localization of TEV P3N-PIPO and its interaction with TEV CI and the intercellular movement-defective mutant CI(AA3,4).
(A) Colocalization of TEV P3N-PIPO-GFP with the PD marker PDLP1-mRFP. Arrows point to the PD-located P3N-PIPO-GFP and PDLP1-mRFP. (B) Coexpression of TEV CI(AA3,4)-mRFP does not change PD-localization of TEV P3N-PIPO-GFP. Arrows indicate PD-located P3N-PIPO-GFP. Arrowheads indicate nucleus-localization of TEV CI(AA3,4)-mRFP. (C) BiFC analysis of interactions of TEV CI-YN and TEV P3N-PIPO-YC (panel I), TEV CI (AA3, 4)-YN and TEV P3N-PIPO-YC(panel II), TEV CI-YN and TEV CI-YC (panel III), and TEV CI (AA3, 4)-YN and TEV CI (AA 3, 4)-YC. N, nucleus. Bars, 10 µm.
Figure 7. Two TEV CI mutants defective in cell-to-cell movement fail to form either cytoplasmic inclusions (when expressed alone) or PD-associated structures (in the presence of P3N-PIPO).
(A) TEV CI-mRFP forms aggregates in the cytoplasm when expressed alone (panel I) and punctate spots along the cell walls when coexpressed with P3N-PIPO-GFP (panel II). (B) When expressed alone, TEV CI(AA3, 4)-mRFP (panel II) and TEV CI(AA100,101)-mRFP (panel III) are distributed in the nucleus and in periphery rather than forming typical inclusions in the cytoplasm (panel I). (C) In the presence of P3N-PIPO, TEV CI(AA3, 4)-mRFP (panel II) and TEV CI(AA100,101)-mRFP (panel III) are distributed in the nucleus and cell periphery rather than targeting PD (panel I). All images are taken 48 hrs post-agroinfiltration. Bars, 8 µm.
Figure 8. Model for potyvirus intercellular transport through PD.
The virion-CI movement complex is intracellularly transported to the modified PD where CI forms conical structures anchored by the PD-located P3N-PIPO. The virion is then fed through the CI structures and PD to enter the adjacent cell. CW, cell wall.
Similar articles
- P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement.
Chai M, Wu X, Liu J, Fang Y, Luan Y, Cui X, Zhou X, Wang A, Cheng X. Chai M, et al. J Virol. 2020 Mar 31;94(8):e01898-19. doi: 10.1128/JVI.01898-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31969439 Free PMC article. - DEVELOPMENTALLY REGULATED PLASMA MEMBRANE PROTEIN of Nicotiana benthamiana contributes to potyvirus movement and transports to plasmodesmata via the early secretory pathway and the actomyosin system.
Geng C, Cong QQ, Li XD, Mou AL, Gao R, Liu JL, Tian YP. Geng C, et al. Plant Physiol. 2015 Feb;167(2):394-410. doi: 10.1104/pp.114.252734. Epub 2014 Dec 24. Plant Physiol. 2015. PMID: 25540331 Free PMC article. - Cell-to-cell movement of plant viruses via plasmodesmata: a current perspective on potyviruses.
Wang A. Wang A. Curr Opin Virol. 2021 Jun;48:10-16. doi: 10.1016/j.coviro.2021.03.002. Epub 2021 Mar 27. Curr Opin Virol. 2021. PMID: 33784579 Review. - Cylindrical Inclusion Protein of Turnip Mosaic Virus Serves as a Docking Point for the Intercellular Movement of Viral Replication Vesicles.
Movahed N, Patarroyo C, Sun J, Vali H, Laliberté JF, Zheng H. Movahed N, et al. Plant Physiol. 2017 Dec;175(4):1732-1744. doi: 10.1104/pp.17.01484. Epub 2017 Oct 31. Plant Physiol. 2017. PMID: 29089395 Free PMC article. - Role of plant virus movement proteins.
Taliansky M, Torrance L, Kalinina NO. Taliansky M, et al. Methods Mol Biol. 2008;451:33-54. doi: 10.1007/978-1-59745-102-4_3. Methods Mol Biol. 2008. PMID: 18370246 Review.
Cited by
- Dynamic Subcellular Localization, Accumulation, and Interactions of Proteins From Tomato Yellow Leaf Curl China Virus and Its Associated Betasatellite.
Li H, Li F, Zhang M, Gong P, Zhou X. Li H, et al. Front Plant Sci. 2020 Jun 16;11:840. doi: 10.3389/fpls.2020.00840. eCollection 2020. Front Plant Sci. 2020. PMID: 32612626 Free PMC article. - Sharka: the past, the present and the future.
Sochor J, Babula P, Adam V, Krska B, Kizek R. Sochor J, et al. Viruses. 2012 Nov 7;4(11):2853-901. doi: 10.3390/v4112853. Viruses. 2012. PMID: 23202508 Free PMC article. Review. - Interaction of the trans-frame potyvirus protein P3N-PIPO with host protein PCaP1 facilitates potyvirus movement.
Vijayapalani P, Maeshima M, Nagasaki-Takekuchi N, Miller WA. Vijayapalani P, et al. PLoS Pathog. 2012;8(4):e1002639. doi: 10.1371/journal.ppat.1002639. Epub 2012 Apr 12. PLoS Pathog. 2012. PMID: 22511869 Free PMC article. - SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection.
Xiong R, Wang A. Xiong R, et al. J Virol. 2013 Apr;87(8):4704-15. doi: 10.1128/JVI.02828-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365455 Free PMC article. - Immunohistochemical localization of Cassava brown streak virus and its morphological effect on cassava leaves.
Saggaf MH, Ndunguru J, Tairo F, Sseruwagi P, Ascencio-Ibáñez JT, Kilalo D, Miano DW. Saggaf MH, et al. Physiol Mol Plant Pathol. 2019 Jan;105:67-76. doi: 10.1016/j.pmpp.2018.06.001. Physiol Mol Plant Pathol. 2019. PMID: 31007375 Free PMC article.
References
- Maule A J. Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol. 2008;11:680–686. - PubMed
- Lucas WJ, Ham BK, Kim J-K. Plasmodesmata-bridging the gap between neiboring cells. Trend Cell Biol. 2009;19:495–503. - PubMed
- Jackson AO, Lim HS, Bragg J, Ganesan U, Lee MY. Hordeivirus replication, movement, and pathogenesis. Annu Rev Phytopathol. 2009;47:385–422. - PubMed
- Verchot-Lubicz J. A new cell-to-cell transport model for Potexviruses. Mol Plant-Microbe Interact. 2005;18:283–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources