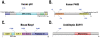GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm - PubMed (original) (raw)
GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm
Yu Xue et al. PLoS One. 2010.
Abstract
As one of the most important and ubiquitous post-translational modifications (PTMs) of proteins, S-nitrosylation plays important roles in a variety of biological processes, including the regulation of cellular dynamics and plasticity. Identification of S-nitrosylated substrates with their exact sites is crucial for understanding the molecular mechanisms of S-nitrosylation. In contrast with labor-intensive and time-consuming experimental approaches, prediction of S-nitrosylation sites using computational methods could provide convenience and increased speed. In this work, we developed a novel software of GPS-SNO 1.0 for the prediction of S-nitrosylation sites. We greatly improved our previously developed algorithm and released the GPS 3.0 algorithm for GPS-SNO. By comparison, the prediction performance of GPS 3.0 algorithm was better than other methods, with an accuracy of 75.80%, a sensitivity of 53.57% and a specificity of 80.14%. As an application of GPS-SNO 1.0, we predicted putative S-nitrosylation sites for hundreds of potentially S-nitrosylated substrates for which the exact S-nitrosylation sites had not been experimentally determined. In this regard, GPS-SNO 1.0 should prove to be a useful tool for experimentalists. The online service and local packages of GPS-SNO were implemented in JAVA and are freely available at: http://sno.biocuckoo.org/.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. The biochemical processes of the endogenous NO source and protein _S_-nitrosylation.
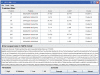
Figure 2. The screen snapshot of GPS-SNO 1.0 software.
The medium threshold was chosen as the default threshold. As an example, the prediction results of human tissue transglutaminase (tTG, P21980) are presented.
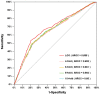
Figure 3. The prediction performance of GPS-SNO 1.0.
The leave-one-out validation and 4-, 6-, 8-, 10-fold cross-validations were calculated. The Receiver Operating Characteristic (ROC) curves and AROCs (area under ROCs) were also carried out.
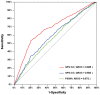
Figure 4. Comparison of GPS 3.0, GPS 2.0 and PSSM.
For comparison, the leave-one-out results of GPS 3.0, GPS 2.0 and PSSM were calculated.
Figure 5. Applications of GPS-SNO 1.0.
Here we predicted potential _S_-nitrosylation sites in experimentally identified _S_-nitrosylated substrates with the default threshold. (A) Human p53 (P04637); (B) Human P4HB (P07237); (C) Mouse Masp1 (P98064); (D) Arabidopsis SAHH1 (O23255).
Similar articles
- GPS-CCD: a novel computational program for the prediction of calpain cleavage sites.
Liu Z, Cao J, Gao X, Ma Q, Ren J, Xue Y. Liu Z, et al. PLoS One. 2011 Apr 20;6(4):e19001. doi: 10.1371/journal.pone.0019001. PLoS One. 2011. PMID: 21533053 Free PMC article. - An efficient support vector machine approach for identifying protein S-nitrosylation sites.
Li YX, Shao YH, Jing L, Deng NY. Li YX, et al. Protein Pept Lett. 2011 Jun;18(6):573-87. doi: 10.2174/092986611795222731. Protein Pept Lett. 2011. PMID: 21271979 - GPS-YNO2: computational prediction of tyrosine nitration sites in proteins.
Liu Z, Cao J, Ma Q, Gao X, Ren J, Xue Y. Liu Z, et al. Mol Biosyst. 2011 Apr;7(4):1197-204. doi: 10.1039/c0mb00279h. Epub 2011 Jan 21. Mol Biosyst. 2011. PMID: 21258675 - Recent Advances in Predicting Protein S-Nitrosylation Sites.
Zhao Q, Ma J, Xie F, Wang Y, Zhang Y, Li H, Sun Y, Wang L, Guo M, Han K. Zhao Q, et al. Biomed Res Int. 2021 Feb 9;2021:5542224. doi: 10.1155/2021/5542224. eCollection 2021. Biomed Res Int. 2021. PMID: 33628788 Free PMC article. Review. - Methodologies for the characterization, identification and quantification of S-nitrosylated proteins.
Foster MW. Foster MW. Biochim Biophys Acta. 2012 Jun;1820(6):675-83. doi: 10.1016/j.bbagen.2011.03.013. Epub 2011 Apr 5. Biochim Biophys Acta. 2012. PMID: 21440604 Free PMC article. Review.
Cited by
- Identification and classification of innexin gene transcripts in the central nervous system of the terrestrial slug Limax valentianus.
Sadamoto H, Takahashi H, Kobayashi S, Kondoh H, Tokumaru H. Sadamoto H, et al. PLoS One. 2021 Apr 15;16(4):e0244902. doi: 10.1371/journal.pone.0244902. eCollection 2021. PLoS One. 2021. PMID: 33857131 Free PMC article. - Proteomic Analysis of _S_-Nitrosation Sites During Somatic Embryogenesis in Brazilian Pine, Araucaria angustifolia (Bertol.) Kuntze.
Borges Araujo AJ, Cerruti GV, Zuccarelli R, Rodriguez Ruiz M, Freschi L, Singh R, Moerschbacher BM, Floh EIS, Wendt Dos Santos AL. Borges Araujo AJ, et al. Front Plant Sci. 2022 Jun 30;13:902068. doi: 10.3389/fpls.2022.902068. eCollection 2022. Front Plant Sci. 2022. PMID: 35845673 Free PMC article. - The SNO-proteome: causation and classifications.
Seth D, Stamler JS. Seth D, et al. Curr Opin Chem Biol. 2011 Feb;15(1):129-36. doi: 10.1016/j.cbpa.2010.10.012. Epub 2010 Nov 17. Curr Opin Chem Biol. 2011. PMID: 21087893 Free PMC article. Review. - CUL4A ubiquitin ligase: a promising drug target for cancer and other human diseases.
Sharma P, Nag A. Sharma P, et al. Open Biol. 2014 Feb 12;4(2):130217. doi: 10.1098/rsob.130217. Open Biol. 2014. PMID: 24522884 Free PMC article. Review. - Prediction of reversible disulfide based on features from local structural signatures.
Sun MA, Wang Y, Zhang Q, Xia Y, Ge W, Guo D. Sun MA, et al. BMC Genomics. 2017 Apr 4;18(1):279. doi: 10.1186/s12864-017-3668-8. BMC Genomics. 2017. PMID: 28376774 Free PMC article.
References
- Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. - PubMed
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. - PubMed
- Tannenbaum SR, White FM. Regulation and specificity of S-nitrosylation and denitrosylation. ACS Chem Biol. 2006;1:615–618. - PubMed
- Hess DT, Matsumoto A, Nudelman R, Stamler JS. S-nitrosylation: spectrum and specificity. Nat Cell Biol. 2001;3:E46–49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous