Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis - PubMed (original) (raw)
Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis
Cristian Bellodi et al. Cancer Res. 2010.
Abstract
Mutations in DKC1, encoding for dyskerin, a pseudouridine synthase that modifies rRNA and regulates telomerase activity, are associated with ribosomal dysfunction and increased cancer susceptibility in the human syndrome, X-linked dyskeratosis congenita (X-DC). In a mouse model for X-DC, impairments in DKC1 function affected the translation of specific mRNAs harboring internal ribosomal entry site (IRES) elements, including the tumor suppressor, p27. However, how this translational deregulation contributes to tumor initiation and progression remains poorly understood. Here, we report that impairment in p27 IRES-mediated translation due to decreased levels of DKC1 activity markedly increases spontaneous pituitary tumorigenesis in p27 heterozygous mice. Using a new bioluminescent mouse model, we monitored p27 translation in vivo and show that p27 IRES-mediated translation is reduced in the pituitary of DKC1 hypomorphic mice (DKC1(m)). Furthermore, we show that DKC1 has a critical role in regulating the assembly of the 48S translational preinitiation complex mediated by the p27 IRES element. An analysis of human tumors identified a novel mutation of DKC1 (DKC1(S485G)) in a human pituitary adenoma. We show that this specific amino acid substitution significantly alters DKC1 stability/pseudouridylation activity, and this correlates with reductions in p27 protein levels. Furthermore, DKC1(S485G) mutation does not alter telomerase RNA levels. Altogether, these findings show that genetic alterations in DKC1 could contribute to tumorigenesis associated with somatic cancers and establish a critical role for DKC1 in tumor suppression, at least in part, through translational control of p27.
(c)2010 AACR.
Figures
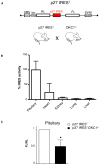
Figure 1. Analysis of p27 IRES-mediated translation using a new bioluminescent mouse model reveal robust translational control in the pituitary, which is reduced in DKC1m background
A) Schematic of the dicistronic transgene used to generate bioluminescent p27 IREST animals. B) Analysis of p27 IREST mice. Graph is mean ± SEM of percentage p27 IRES activity (FLuc values) measured in different organs prepared from three p27 IREST mice and normalized by the mRNA dicistronic expression. C) p27 IRES-mediated translation is markedly reduced in DKC1m pituitary glands. Levels of p27 IRES-mediated translation were measured in pituitary glands isolated from p27 IREST and p27 IREST;DKC1m mice. Columns, mean ± SEM of ratio FLuc/RLuc (FL/RL) measured in four pairs of age-matched animals; bars, SEM. Statistical analysis was carried out using Student t test. *P<0.05.
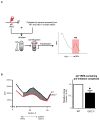
Figure 2. Molecular mechanism of p27 IRES-mediated translation impairment in DKC1m cells
A) Schematic of the biochemical approach used to study the 48S complex formation in WT and DKC1m MEFs. A [32P]-p27 IRES mRNA probe was incubated with cytoplasmic extracts. Newly assembled 48S complexes were separated by sucrose gradient centrifugation and fractionated according to their density. A peak of radioactivity was generated and coincided with fractions containing the 48S complexes. B) Representative of the 48S subunit quantification from total cytoplasmic extracts prepared from serum starved (0.1% FBS) WT and DKC1m cells. Representative profile of a sucrose density gradient reporting the radioactive intensity per fractions in WT (black) and DKC1m (red) extracts, respectively (left). Columns, mean ± SEM of the area under the curve in WT and DKC1m cell extracts measured in 3 independent experiments; bars, SEM (right). Statistical analysis was carried out using Student t test. *P<0.05.
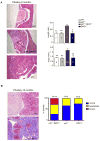
Figure 3. DKC1 and p27 genetically cooperate toward pituitary tumorigenesis
A) Micrographs of hematoxylin and eosin-stained paraffin sections prepared from pituitary glands of 8 month old WT, p27+/−;DKC1m (hyperplasia) and p27−/− (tumor) animals. Numbers on the pictures indicate: (1) pars nervosa, (2) pars intermedia (intermediate lobe) and (3) pars distalis, and bars correspond to 10 nm. Columns, mean ± SEM of length and area of the intermediate lobe; bars SEM. The number of animals analyzed for each genotype is indicated on the columns. Statistical analysis was performed using unpaired two-tailed t test. P values are ** <0.01 and * <0.05. B) Representative histopathological analysis of a normal pituitary gland (top) and a pituitary adenoma (bottom) from 15 month old WT and p27+/−;DKC1m mice, respectively. Graph shows the percentages of double p27+/−;DKC1m, p27+/− and DKC1m animals with a normal pituitary (blue), pituitary hyperplasia (yellow) and pituitary cancers (red).
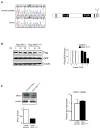
Figure 4. Identification of a novel DKC1 mutation in a human pituitary adenoma
A) Chromatographs showing a novel mutation in the DKC1 locus identified in a human pituitary adenoma (left). Scheme of the mutation, an A1493G variation located on one DKC1 allele leads to the amino acid change S485G in the C-terminal portion of the protein (right). B) Analysis of DKC1S485G protein stability. Densitometric analysis of the DKC1 levels was done at each time point after blockage of protein synthesis as indicated. GFP was co-transfected for transfection efficiency and β-actin was used as loading control. C) DKC1 protein (left) and mRNA (right) levels in a normal pituitary and in the tumor bearing the mutation DKC1S485G. Densitometric analysis of DKC1 over β-actin protein levels in each sample is shown (bottom left).
Figure 5. Impaired rRNA pseudouridylation is associated with reductions of p27 protein levels but not of the telomerase RNA component (TERC) in the DKC1S485G mutant tumor
A) Chromatographs of HPLC analysis of the 18S rRNA pseudouridylation are shown. Bar graph shows quantification of the Ψ/C ratio in samples from normal pituitary and the DKC1S485G mutant tumor. B) The levels of the TERC were measured by QPCR in two normal pituitaries and the DKC1S485G pituitary tumor. Analysis was performed in triplicate on two different samples per specimen. No significant differences between the samples were found. _P_>0.05 was determined employing the unpaired two-tailed t test. C) p27 protein levels are markedly reduced in the tumor carrying the DKC1S485G mutation. Western blot (left) and QPCR analysis (right) of p27 expression was performed using protein extracts and total RNA prepared from a normal pituitary and the tumor carrying the DKC1S485G mutantion. Densitometric analysis of p27 over β-actin is shown (bottom left). No significant differences in p27 mRNA levels were measured between the samples. _P_>0.05 was determined employing the unpaired two-tailed t test.
Similar articles
- Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita.
Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Yoon A, et al. Science. 2006 May 12;312(5775):902-6. doi: 10.1126/science.1123835. Science. 2006. PMID: 16690864 - P27/CDKN1B Translational Regulators in Pituitary Tumorigenesis.
Martins CS, Camargo RC, Saggioro FP, Neder L, Machado HR, Moreira AC, de Castro M. Martins CS, et al. Horm Metab Res. 2016 Dec;48(12):840-846. doi: 10.1055/s-0042-118613. Epub 2016 Nov 7. Horm Metab Res. 2016. PMID: 27824399 - Severity of X-linked dyskeratosis congenita (DKCX) cellular defects is not directly related to dyskerin (DKC1) activity in ribosomal RNA biogenesis or mRNA translation.
Thumati NR, Zeng XL, Au HH, Jang CJ, Jan E, Wong JM. Thumati NR, et al. Hum Mutat. 2013 Dec;34(12):1698-707. doi: 10.1002/humu.22447. Epub 2013 Oct 21. Hum Mutat. 2013. PMID: 24115260 - Tumor suppressor loss in pituitary tumors.
Alexander JM. Alexander JM. Brain Pathol. 2001 Jul;11(3):342-55. doi: 10.1111/j.1750-3639.2001.tb00404.x. Brain Pathol. 2001. PMID: 11414476 Free PMC article. Review. - Dyskeratosis congenita: molecular insights into telomerase function, ageing and cancer.
Marrone A, Dokal I. Marrone A, et al. Expert Rev Mol Med. 2004 Dec 20;6(26):1-23. doi: 10.1017/S1462399404008671. Expert Rev Mol Med. 2004. PMID: 15613268 Review.
Cited by
- A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs.
Taoka M, Nobe Y, Hori M, Takeuchi A, Masaki S, Yamauchi Y, Nakayama H, Takahashi N, Isobe T. Taoka M, et al. Nucleic Acids Res. 2015 Oct 15;43(18):e115. doi: 10.1093/nar/gkv560. Epub 2015 May 26. Nucleic Acids Res. 2015. PMID: 26013808 Free PMC article. - Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis.
Liu B, Zhang J, Huang C, Liu H. Liu B, et al. PLoS One. 2012;7(8):e43147. doi: 10.1371/journal.pone.0043147. Epub 2012 Aug 13. PLoS One. 2012. PMID: 22912812 Free PMC article. - Evaluation of dyskerin expression and the Cajal body protein WRAP53β as potential prognostic markers for patients with primary vaginal carcinoma.
Ranhem C, Larsson GL, Lindqvist D, Sorbe B, Karlsson MG, Farnebo M, Hellman K, Kovaleska L, Kashuba E, Andersson S. Ranhem C, et al. Oncol Lett. 2022 Jan;23(1):30. doi: 10.3892/ol.2021.13148. Epub 2021 Nov 23. Oncol Lett. 2022. PMID: 34868367 Free PMC article. - Epitranscriptomics of cancer.
Tusup M, Kundig T, Pascolo S. Tusup M, et al. World J Clin Oncol. 2018 Jun 10;9(3):42-55. doi: 10.5306/wjco.v9.i3.42. World J Clin Oncol. 2018. PMID: 29900123 Free PMC article. Review.
References
- Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73:103–12. - PubMed
- Montanaro L, Brigotti M, Clohessy J, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J Pathol. 2006;210:10–8. - PubMed
- Poncet D, Belleville A, t’kint de Roodenbeke C, et al. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111:2388–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL085572/HL/NHLBI NIH HHS/United States
- 3R01HL085572-05S1/HL/NHLBI NIH HHS/United States
- R01 HL085572-05/HL/NHLBI NIH HHS/United States
- R01 HL085572-05S1/HL/NHLBI NIH HHS/United States
- R01 HL085572-04/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases