Mammalian evolution may not be strictly bifurcating - PubMed (original) (raw)
Mammalian evolution may not be strictly bifurcating
Björn M Hallström et al. Mol Biol Evol. 2010 Dec.
Abstract
The massive amount of genomic sequence data that is now available for analyzing evolutionary relationships among 31 placental mammals reduces the stochastic error in phylogenetic analyses to virtually zero. One would expect that this would make it possible to finally resolve controversial branches in the placental mammalian tree. We analyzed a 2,863,797 nucleotide-long alignment (3,364 genes) from 31 placental mammals for reconstructing their evolution. Most placental mammalian relationships were resolved, and a consensus of their evolution is emerging. However, certain branches remain difficult or virtually impossible to resolve. These branches are characterized by short divergence times in the order of 1-4 million years. Computer simulations based on parameters from the real data show that as little as about 12,500 amino acid sites could be sufficient to confidently resolve short branches as old as about 90 million years ago (Ma). Thus, the amount of sequence data should no longer be a limiting factor in resolving the relationships among placental mammals. The timing of the early radiation of placental mammals coincides with a period of climate warming some 100-80 Ma and with continental fragmentation. These global processes may have triggered the rapid diversification of placental mammals. However, the rapid radiations of certain mammalian groups complicate phylogenetic analyses, possibly due to incomplete lineage sorting and introgression. These speciation-related processes led to a mosaic genome and conflicting phylogenetic signals. Split network methods are ideal for visualizing these problematic branches and can therefore depict data conflict and possibly the true evolutionary history better than strictly bifurcating trees. Given the timing of tectonics, of placental mammalian divergences, and the fossil record, a Laurasian rather than Gondwanan origin of placental mammals seems the most parsimonious explanation.
Figures
FIG. 1.
Presence/absence matrix for illustrating sequence density. Each dash indicates the presence of a given gene (x axis) in a given species (y axis). The matrix is sorted for maximum sequence density along both axes, placing the species with the best coverage at the top of the plot and the genes represented by the most species at the left.
FIG. 2.
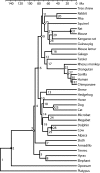
ML tree reconstructed from an alignment length of 954 ka aa from 37 species under the WAG2000 + 8GI of sequence evolution.
FIG. 3.
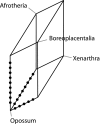
Chronogram of mammalian divergences. Open and filled circles indicate calibration points: Open circles represent those estimated divergence times anywhere between the boundaries, filled circles those reaching either the upper or lower boundary.
FIG. 4.
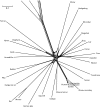
Neighbor-net based on the 954 ka aa alignment from 37 species. All intraordinal, ordinal, and most superordinal relationships are clearly defined in the neighbor-net by stretched boxes that are longer than they are wide, indicating limited conflict in the data. The clades that are poorly supported by ML aa sequence data analysis are characterized by boxed nodes that are nearly square or by negligibly short branch lengths.
FIG. 5.
Close up of the neighbor-net highlighting major splits.
FIG. 6.
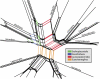
Retroposon split network.
FIG. 7.
Simulation of aa sequence lengths required for resolving temporally tight divergences at 90 Ma. The black line is reconstructed from the simulation of a data set free of gaps, whereas the gray line represents a simulation where gaps were included according to their frequency in the original data.
Similar articles
- Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations.
Hallström BM, Janke A. Hallström BM, et al. BMC Evol Biol. 2008 May 27;8:162. doi: 10.1186/1471-2148-8-162. BMC Evol Biol. 2008. PMID: 18505555 Free PMC article. - The Interrelationships of Placental Mammals and the Limits of Phylogenetic Inference.
Tarver JE, Dos Reis M, Mirarab S, Moran RJ, Parker S, O'Reilly JE, King BL, O'Connell MJ, Asher RJ, Warnow T, Peterson KJ, Donoghue PC, Pisani D. Tarver JE, et al. Genome Biol Evol. 2016 Jan 5;8(2):330-44. doi: 10.1093/gbe/evv261. Genome Biol Evol. 2016. PMID: 26733575 Free PMC article. - A genomic approach to examine the complex evolution of laurasiatherian mammals.
Hallström BM, Schneider A, Zoller S, Janke A. Hallström BM, et al. PLoS One. 2011;6(12):e28199. doi: 10.1371/journal.pone.0028199. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164244 Free PMC article. - Mammal madness: is the mammal tree of life not yet resolved?
Foley NM, Springer MS, Teeling EC. Foley NM, et al. Philos Trans R Soc Lond B Biol Sci. 2016 Jul 19;371(1699):20150140. doi: 10.1098/rstb.2015.0140. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27325836 Free PMC article. Review. - Mammalian evolution and biomedicine: new views from phylogeny.
Springer MS, Murphy WJ. Springer MS, et al. Biol Rev Camb Philos Soc. 2007 Aug;82(3):375-92. doi: 10.1111/j.1469-185X.2007.00016.x. Biol Rev Camb Philos Soc. 2007. PMID: 17624960 Review.
Cited by
- Genome of the Chinese tree shrew.
Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, He J, Hou HL, Hu L, Hu XT, Jiang XT, Lai R, Lang YS, Liang B, Liao SG, Mu D, Ma YY, Niu YY, Sun XQ, Xia JQ, Xiao J, Xiong ZQ, Xu L, Yang L, Zhang Y, Zhao W, Zhao XD, Zheng YT, Zhou JM, Zhu YB, Zhang GJ, Wang J, Yao YG. Fan Y, et al. Nat Commun. 2013;4:1426. doi: 10.1038/ncomms2416. Nat Commun. 2013. PMID: 23385571 - Reticulation, divergence, and the phylogeography-phylogenetics continuum.
Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C. Edwards SV, et al. Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):8025-32. doi: 10.1073/pnas.1601066113. Proc Natl Acad Sci U S A. 2016. PMID: 27432956 Free PMC article. - Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the laurasiatherian mammals.
Zhou X, Xu S, Xu J, Chen B, Zhou K, Yang G. Zhou X, et al. Syst Biol. 2012 Jan;61(1):150-64. doi: 10.1093/sysbio/syr089. Epub 2011 Sep 7. Syst Biol. 2012. PMID: 21900649 Free PMC article. - Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference.
Segura J, Ferretti L, Ramos-Onsins S, Capilla L, Farré M, Reis F, Oliver-Bonet M, Fernández-Bellón H, Garcia F, Garcia-Caldés M, Robinson TJ, Ruiz-Herrera A. Segura J, et al. Proc Biol Sci. 2013 Sep 25;280(1771):20131945. doi: 10.1098/rspb.2013.1945. Print 2013 Nov 22. Proc Biol Sci. 2013. PMID: 24068360 Free PMC article. - Conventional simulation of biological sequences leads to a biased assessment of multi-Loci phylogenetic analysis.
Aguiar BO, Schrago CG. Aguiar BO, et al. Evol Bioinform Online. 2013 Aug 13;9:317-25. doi: 10.4137/EBO.S12483. eCollection 2013. Evol Bioinform Online. 2013. PMID: 23997573 Free PMC article.
References
- Archibald JD. Timing and biogeography of the eutherian radiation: fossils and molecules compared. Mol Phylogenet Evol. 2003;28:350–359. - PubMed
- Arnason U, Adegoke JA, Gullberg A, Harley EH, Janke A, Kullberg M. Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene. 2008;421:37–51. - PubMed
- Arnason U, Gullberg A, Gretarsdottir S, Ursing B, Janke A. The mitochondrial genome of the sperm whale and a new molecular reference for estimating eutherian divergence dates. J Mol Evol. 2000;50:569–578. - PubMed
- Asher RJ. Insectivoran-grade placental mammals: character evolution and fossil history. In: Rose KD, Archibald JD, editors. The rise of placental mammals. Baltimore (MD): The Johns Hopkins University Press; 2005. pp. 50–70.
- Asher RJ, Novacek MJ, Geisler JH. Relationships of endemic African mammals and their fossil relatives based on morphological and molecular evidence. J Mamm Evol. 2003;10:131–162.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources