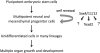Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors - PubMed (original) (raw)
Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors
Pallavi Bhattaram et al. Nat Commun. 2010.
Free PMC article
Abstract
During organogenesis, neural and mesenchymal progenitor cells give rise to many cell lineages, but their molecular requirements for self-renewal and lineage decisions are incompletely understood. In this study, we show that their survival critically relies on the redundantly acting SoxC transcription factors Sox4, Sox11 and Sox12. The more SoxC alleles that are deleted in mouse embryos, the more severe and widespread organ hypoplasia is. SoxC triple-null embryos die at midgestation unturned and tiny, with normal patterning and lineage specification, but with massively dying neural and mesenchymal progenitor cells. Specific inactivation of SoxC genes in neural and mesenchymal cells leads to selective apoptosis of these cells, suggesting SoxC cell-autonomous roles. Tead2 functionally interacts with SoxC genes in embryonic development, and is a direct target of SoxC proteins. SoxC genes therefore ensure neural and mesenchymal progenitor cell survival, and function in part by activating this transcriptional mediator of the Hippo signalling pathway.
Figures
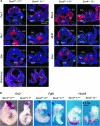
Figure 1. SoxC genes functionally interact with each other in multiple developmental processes.
(a) E18.5 Sox4 +/− 11 +/− and Sox11 −/− littermates often display omphalocele (arrows), and Sox11 −/− mice have open eyelids and cleft lips. (b) E18.5 Sox11 −/− mice have a common arterial trunk (CT), and Sox4 +/− 11 +/− mice have the arterial (AT) and pulmonary (PT) trunks emerging from the right ventricle (RV). LV, left ventricle. (c) E18.5 fetus skeletal preparations, in which non-mineralized cartilage is stained with Alcian blue and mineralized bone and cartilage with Alizarin red, reveal that vertebral bodies (arrows, shown from the third lumbar vertebra—L3—to the second sacral vertebra—S2) are well formed in control mice, but that L4 and L5 are duplicated in Sox11 −/− and Sox4 +/− 11 +/− mice. (d) Skeletal preparations of the thoracic cage show that the five sternebrae (numbers 1–5) and the xyphoid process (X) are mineralized and distinct in control mice, whereas the last two or three sternebrae and the xyphoid process of Sox11 −/− and Sox4 +/− 11 +/− mice are misshapen and irregularly mineralized. (e) E18.5 Sox4 +/− , Sox4 +/− 11 +/− and Sox4 +/− 11 +/− 12 −/− littermates look normal. (f) Sox4 +/− and Sox4 +/− 11 +/− fetuses have distinct aortic and pulmonary trunks, but Sox4 +/− 11 +/− 12 −/− littermates have a common trunk. (g) Several Sox4 +/− 11 +/− 12 −/− lumbar vertebral bodies are fused (double arrowheads) along the antero-posterior axis, in addition to being duplicated (arrows) along the midline. (h) Skeletal preparations show that the sternal bars of Sox4 +/− 11 +/− 12 −/− embryos are barely fused at the midline (arrow) and that the xyphoid process (X) and fifth (5) sternebra are hardly, if at all, mineralized.
Figure 2. SoxC genes are necessary in early organogenesis.
(a) Side views of E9.5 Sox4 −/− 11 +/−, Sox4 −/− 11 +/− and Sox4 −/− 11 −/− littermates show that the latter two types of mutants are smaller and not fully turned. fb, forebrain; asterisk (*), branchial arches. (b) Scanning electron microscopy of E9.5 embryos. The top row shows ventral views of the head and heart, and the bottom row shows dorsal views. Note the lack of heart (h) coiling in the Sox4 −/− 11 +/− embryo, cardia bifida in the Sox4 −/− 11 −/− embryo and incomplete closure of the neural tube in the cephalic region (arrows) in Sox4 −/− 11 +/− and Sox4 −/− 11 −/− embryos. lb, limb bud. (c) Histology sections of E9.5 embryos made as indicated by the schematics and stained with haematoxylin and eosin show that the Sox4 −/− 11 +/− and Sox4 −/− 11 −/− branchial arch mesenchyme (b) is loose and small. The Sox4 −/− 11 −/− neural tube (n) is thin and wavy. Somites (s) are small. Heart primordia (h) are not fused. Sox4 −/− 11 +/− embryos are partially turned, and Sox4 −/− 11 −/− embryos are not turned. (d) Histology sections show that the neural tube (n) is thinner and wavier in E9.5 Sox4 −/− 11 −/− 12 −/− embryos than in Sox4 −/− 11 −/− littermates, whereas the first branchial arch (b) mesenchyme is similar in size and degree of cellularity in the two types of embryos.
Figure 3. Embryo axial patterning and tissue specification are not affected by SoxC inactivation.
(a) RNA in situ hybridizations of E9.5 embryo sections demonstrate that Pax7 (dorsal neural tube and dermomyotome), Ptc1 (ventral neural tube, paraxial mesoderm, splanchnic mesoderm), Shh (notochord and neural floor plate), Brachyury (notochord), Bmp4 (lateral/ventral mesoderm), Myf5 (myotome) and Pax1 (sclerotome) are expressed normally in Sox4 −/− 11 −/− embryos. lp, lateral plate mesoderm; n, neural tube; so, somite, sm, splanchnic mesoderm. RNA signals are seen in red and cell nuclei are stained in blue with the Hoechst dye 33258. (b) Whole-mount RNA in situ hybridizations show that the expression domains of Otx2 (forebrain), Fgf8 (midbrain, arrows) and Hoxb9 (spinal cord) are maintained in E9.5 Sox4 −/− 11 −/− embryos.
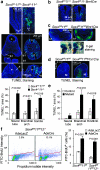
Figure 4. SoxC genes are required for neural and mesenchymal cell survival.
(a) TdT-mediated dUTP nick end labeling (TUNEL) assay in E9.5 embryo sections shows increased cell death in the neural tube (n), first branchial arch mesenchyme (b1) and somites (s) of Sox4 −/− 11 −/− embryos compared with Sox4 +/− 11 +/− littermates. Cell nuclei are rendered blue with 4,6-diamidino-2-phenylindole (DAPI), and apoptotic bodies fluoresce in green. The percentage of TUNEL versus DAPI signal in areas of interest is shown as average with standard deviation obtained for three sections in each of three embryos per genotype. Statistical significance was determined using a Student's two-tailed, paired _t-_test. (b) TUNEL assay in embryo sections shows increased cell death in the neural tube (n) of E10.5 Sox4 fl/fl 11 −/− Brn4Cre embryos. (c) TUNEL assay in embryo sections shows increased cell death in the branchial arch mesenchyme (b1) of E10.5 Sox4 fl/fl 11 fl/fl Wnt1Cre embryos. 5-bromo-4-chloro-3-indolyl-β-
D
-galactoside staining of Sox4 fl/fl 11 fl/fl and Sox4 fl/fl 11 fl/fl R26 lacZ Wnt1Cre littermates shows that neural crest cells have migrated to proper sites in the first (b1) and second (b2) branchial arches of the mutants. (d) TUNEL assay in embryo sections shows increased cell death in the hindlimb mesenchyme (hl) of E11.5 Sox4 fl/fl 11 fl/fl Prx1Cre embryos. The antero-posterior (A→P) and proximo–distal (P→D) axes of limb buds are indicated. (e) Percentage of TUNEL-positive cells or area in control and mutant tissues of interest. Quantification was carried out as described in panel a. (f) Flow cytometry TUNEL assay shows more dead cells in Sox4 fl/fl 11 fl/fl and Sox4 fl/fl 11 fl/fl 12 −/− primary limb bud cell cultures treated with Cre-expressing adenovirus (AdeCre) than in AdeLacZ-treated control cultures. Left panels, fluorescence-activated cell sorting profiles of representative samples. The percentage of dead cells (BrdU+, above the red line) is indicated. Right panel, average with standard deviation of data obtained from three independent experiments. Statistical significance was determined using a Student's two-tailed, paired _t_-test.
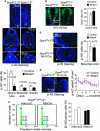
Figure 5. The rate of cell proliferation is reduced in Sox4 −/− 11 −/− embryos, not in conditional mutants.
(a) E9.5 embryo sections immunostained for phospho-histone-3 (p-H3, green signal) and counterstained with 4,6-diamidino-2-phenylindole (blue signal) show a lower rate of cell proliferation in the subventricular layer of the neural tube (n), first branchial arch (b1) and somitic mesenchyme (s) of Sox4 −/− 11 −/− embryos than in Sox4 +/− 11 +/− littermates. The percentage of p-H3-positive cells is shown as the average with standard deviation for three sections in each of three embryos per genotype. Statistical significance was determined using a Student's two-tailed, paired t test. (b) Immunostaining of BrdU incorporation (green signal) in E10.5 Sox4 fl/fl 11 +/+ and Sox4 fl/fl 11 −/− Brn4Cre littermates. Quantification was carried out as described in a. (c) p-H3 staining of sections from E10.5 Sox4 fl/fl 11 fl/fl and Sox4 fl/fl 11 fl/fl R26 lacZ Wnt1Cre littermates at the level of the first branchial arches (b1). Quantification was carried out as described in a. (d) p-H3 staining of hindlimb bud sections from E11.5 Sox4 fl/fl 11 fl/fl and Sox4 fl/fl 11 fl/fl Prx1Cre littermates. The number of p-H3-positive cells was calculated per 0.01 mm2 in up to 11 consecutive layers drawn along the proximo–distal axis (P→D). The plot shows the averages with standard deviation of p-H3-positive cells per surface area in three adjacent sections for a representative experiment. (e) Analysis of cell-cycle distribution by propidium iodide staining of primary limb bud cells from E11.5 Sox4 fl/fl 11 fl/fl 12 −/− embryos 24 h after infection with AdeLacZ or AdeCre. Representative fluorescence-activated cell sorting profiles are shown. The percentages of cells in S and G2 phase are shown for two independent experiments.
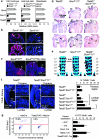
Figure 6. Tead2 is expressed and functions downstream of the SoxC genes.
(a) Quantitative real-time reverse transcription–PCR shows RNA levels of Sox4, Sox11 and Tead2 relative to Gapdh in control and mutant samples. Data are presented as average with standard deviation from three independent samples. Control and mutant data are statistically different (P<0.002, two-tailed Student's _t_-test). (b) RNA in situ hybridization of E9.5 embryo sections shows Tead2 downregulation in Sox4 −/− 11 −/− neural tissue (nt) and somites (s). (c) RNA in situ hybridization shows Tead2 downregulation in limb bud mesenchyme (lb) of E10.5 Sox4 fl/fl 11 fl/fl Prx1Cre mutants, but not in neural tube (nt) and other sites. (d) Frontal sections show a normal heart in E18.5 Tead2 −/− fetuses. One Sox4 +/− 11 +/− embryo (out of 4: _n_=4) and two Sox4 +/− 11 +/− Tead2 +/− embryos (_n_=9) had ventricular septation defect. All Tead2 −/− Sox4 +/− 11 +/− littermates (_n_=7) had this defect (arrow) and arterial (AT) and pulmonary (PT) trunks arising from the right ventricle. The septation defect is more frequent in Sox4 +/− 11 +/− Tead2 −/− than in Sox4 +/− 11 +/− and Sox4 +/− 11 +/− Tead2 +/− fetuses (_P_=0.024 and 0.003, respectively, two-tailed, Fisher's exact test). (e) Skeletal preparations show that all Tead2 −/− fetuses (_n_=8) had normal vertebral bodies (arrows). All Sox4 +/− 11 +/− (_n_=4), Sox4 +/− 11 +/− Tead2 +/− (_n_=9) and Sox4 +/− 11 +/− Tead2 −/− littermates (_n_=7) had duplication of several lumbar vertebral bodies, and six Sox4 +/− 11 +/− Tead2 −/− fetuses also had vertebral body antero-posterior fusions (double arrowheads). The latter phenotype was significantly different from that of littermates (_P_≤0.015, two-tailed, Fisher's exact test). (f) TdT-mediated dUTP nick end labeling assay in E11.5 embryo sections shows increased cell death around the notochord in the lumbar, but not in the thoracic region, in Sox4 +/− 11 +/− Tead2 −/− embryos compared with control littermates. Left, representative pictures. Right, average with standard deviation of data from three embryos per genotype. Brackets link statistically different data (_P_-value <0.01, Student's two-tailed, paired _t_-test). (g) Apoptosis assay in Sox4 fl/fl 11 fl/fl and Sox4 fl/fl 11 fl/fl 12 −/− osteoblasts. Cells were treated with AdeLacZ or AdeCre after transfection with expression plasmids for Amcyan protein and no protein (−), Sox4, Tead2 or Tead2/YAP. Left, representative fluorescence-activated cell sorting profiles. The percentage of Amcyan+ dying cells is indicated. Right, average with standard deviation of data from three independent experiments. Sox4,Cre and Tead2/YAP,Cre data are statistically different from [−,Cre] data (P<0.01, two-tailed Student's _t_-test).
Figure 7. SoxC proteins directly activate Tead2.
(a) Schematic of the mouse Tead2 locus from upstream of the transcription start site (angled arrow) to downstream of the translation start site (ATG) in the second exon, and analysis of mammalian conservation. Highly conserved, non-coding sequences were found in the promoter and first exon (PE1 box) and in the first intron (InS box). No other highly conserved region was found in other Tead2 introns or in the agenic region around Tead2 (data not shown). Arrows, position of primers used in chromatin immunoprecipitation assay. Numbers 1–4 indicate the position of Sox-binding sites. (b) Tead2 reporters were made by cloning the indicated Tead2 sequences upstream of the firefly luciferase gene. (c) Transient transfection of Cos-1 cells with Tead2 reporters and expression vectors for Sox4, Sox11 or no protein (−). Reporter activities (RLU, relative luciferase units) were normalized for transfection efficiency and are presented as the average with standard deviation of data obtained from three independent cultures. Constructs bearing PE1+InS are significantly activated by SoxC proteins (P<0.001, Student's _t_-test), unless the SoxC-binding sites are mutated (PE1+InS-Mut). (d) ClustalW alignment of sequences encompassing putative SoxC-binding sites in several mammalian genomes. These sites are boxed. These sequences and mutant versions, as indicated, were used to generate electrophoretic mobility shift assay probes. (e) Electrophoretic mobility shift assay with Tead2 probes and protein extracts from Cos-1 cells transfected with a Sox4 expression plasmid. Sox4 binds to all wild-type probes (arrowhead), but not to mutant probes, except m1b. (f) Chromatin immunoprecipitation assay of SoxC proteins binding to endogenous Tead2. Chromatin extracts from C3H-10T1/2 cells transiently expressing FLAG-Sox proteins were precipitated in the absence (no IgG) or presence of non-immune (n.i.) rabbit IgG, RNA polymerase II antibody or FLAG antibody. PCR products from immunoprecipitated chromatin and input are shown on resolution by agarose gel electrophoresis. DNA markers and the size of PCR products are shown.
Figure 8. Model for the roles of SoxC genes in organogenesis.
Organogenesis starts as pluripotent embryonic stem cells develop into multipotent neural and mesenchymal cells. These progenitor cells are important for embryo growth and organogenesis through their ability to self-renew and give rise to many different cell types. This study demonstrated that SoxC proteins—Sox4, Sox11 and Sox12—have a primary role in ensuring the survival of these cells. They directly activate the gene for Tead2, a transcriptional mediator of the Hippo signalling pathway that is capable of mediating at least some of their cell survival functions. It is likely, however, that they also fulfil some of their functions through the activation of other genes that remain to be uncovered.
Similar articles
- The transcription factor protein Sox11 enhances early osteoblast differentiation by facilitating proliferation and the survival of mesenchymal and osteoblast progenitors.
Gadi J, Jung SH, Lee MJ, Jami A, Ruthala K, Kim KM, Cho NH, Jung HS, Kim CH, Lim SK. Gadi J, et al. J Biol Chem. 2013 Aug 30;288(35):25400-25413. doi: 10.1074/jbc.M112.413377. Epub 2013 Jul 25. J Biol Chem. 2013. PMID: 23888050 Free PMC article. - Orchestration of Neuronal Differentiation and Progenitor Pool Expansion in the Developing Cortex by SoxC Genes.
Chen C, Lee GA, Pourmorady A, Sock E, Donoghue MJ. Chen C, et al. J Neurosci. 2015 Jul 22;35(29):10629-42. doi: 10.1523/JNEUROSCI.1663-15.2015. J Neurosci. 2015. PMID: 26203155 Free PMC article. - SOXC are critical regulators of adult bone mass.
Angelozzi M, Karvande A, Lefebvre V. Angelozzi M, et al. Nat Commun. 2024 Apr 5;15(1):2956. doi: 10.1038/s41467-024-47413-2. Nat Commun. 2024. PMID: 38580651 Free PMC article. - Critical roles for SoxC transcription factors in development and cancer.
Penzo-Méndez AI. Penzo-Méndez AI. Int J Biochem Cell Biol. 2010 Mar;42(3):425-8. doi: 10.1016/j.biocel.2009.07.018. Epub 2009 Aug 3. Int J Biochem Cell Biol. 2010. PMID: 19651233 Free PMC article. Review. - SOXC Genes and the Control of Skeletogenesis.
Lefebvre V, Bhattaram P. Lefebvre V, et al. Curr Osteoporos Rep. 2016 Feb;14(1):32-8. doi: 10.1007/s11914-016-0296-1. Curr Osteoporos Rep. 2016. PMID: 26830765 Free PMC article. Review.
Cited by
- Phosphorylation of the neurogenic transcription factor SOX11 on serine 133 modulates neuronal morphogenesis.
Balta EA, Schäffner I, Wittmann MT, Sock E, von Zweydorf F, von Wittgenstein J, Steib K, Heim B, Kremmer E, Häberle BM, Ueffing M, Lie DC, Gloeckner CJ. Balta EA, et al. Sci Rep. 2018 Nov 1;8(1):16196. doi: 10.1038/s41598-018-34480-x. Sci Rep. 2018. PMID: 30385877 Free PMC article. - SOXopathies: Growing Family of Developmental Disorders Due to SOX Mutations.
Angelozzi M, Lefebvre V. Angelozzi M, et al. Trends Genet. 2019 Sep;35(9):658-671. doi: 10.1016/j.tig.2019.06.003. Epub 2019 Jul 6. Trends Genet. 2019. PMID: 31288943 Free PMC article. Review. - The RNA exosome nuclease complex regulates human embryonic stem cell differentiation.
Belair C, Sim S, Kim KY, Tanaka Y, Park IH; and; Wolin SL. Belair C, et al. J Cell Biol. 2019 Aug 5;218(8):2564-2582. doi: 10.1083/jcb.201811148. Epub 2019 Jul 15. J Cell Biol. 2019. PMID: 31308215 Free PMC article. - Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-Resistant Prostate Cancer.
Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, Le Magnen C, Chester D, Mostaghel EA, Califano A, Rubin MA, Shen MM, Abate-Shen C. Zou M, et al. Cancer Discov. 2017 Jul;7(7):736-749. doi: 10.1158/2159-8290.CD-16-1174. Epub 2017 Apr 14. Cancer Discov. 2017. PMID: 28411207 Free PMC article. - Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization.
Chiang SY, Wu HC, Lin SY, Chen HY, Wang CF, Yeh NH, Shih JH, Huang YS, Kuo HC, Chou SJ, Chen RH. Chiang SY, et al. Sci Adv. 2021 Feb 12;7(7):eabc6093. doi: 10.1126/sciadv.abc6093. Print 2021 Feb. Sci Adv. 2021. PMID: 33579706 Free PMC article.
References
- Bernardo M. E., Locatelli F. & Fibbe W. E. Mesenchymal stromal cells. Ann. NY Acad. Sci. 1176, 101–117 (2009). - PubMed
- Kuhn N. Z. & Tuan R. S. Regulation of stemness and stem cell niche of mesenchymal stem cells: implications in tumorigenesis and metastasis. J. Cell. Physiol. 222, 268–277 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases