Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3 - PubMed (original) (raw)
Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3
Jacob S Yount et al. Nat Chem Biol. 2010 Aug.
Abstract
Identification of immune effectors and the post-translational modifications that control their activity is essential for dissecting mechanisms of immunity. Here we demonstrate that the antiviral activity of interferon-induced transmembrane protein 3 (IFITM3) is post-translationally regulated by S-palmitoylation. Large-scale profiling of palmitoylated proteins in a dendritic cell line using a chemical reporter strategy revealed over 150 lipid-modified proteins with diverse cellular functions, including innate immunity. We discovered that S-palmitoylation of IFITM3 on membrane-proximal cysteines controls its clustering in membrane compartments and its antiviral activity against influenza virus. The sites of S-palmitoylation are highly conserved among the IFITM family of proteins in vertebrates, which suggests that S-palmitoylation of these immune effectors may be an ancient post-translational modification that is crucial for host resistance to viral infections. The S-palmitoylation and clustering of IFITM3 will be important for elucidating its mechanism of action and for the design of antiviral therapeutics.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
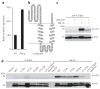
Figure 1. Visualization and identification of palmitoylated proteins in DC2.4 cells
(a) Metabolic labeling of cells with alk-16 palmitate reporter and subsequent CuAAC ligation with bioorthogonal detection tags for imaging or proteomics. (b,c) DC2.4 cells were incubated for 2 h with 50 mM alk-16 or DMSO as a control. In b, cell lysates were reacted with az-rho by CuACC, and proteins were separated by SDS-PAGE for visualization by fluorescence gel scanning. Coomassie blue staining demonstrates comparable loading. In c, cell lysates were reacted with az-diazo-biotin by CuAAC for enrichment of alk-16–labeled proteins with streptavidin beads and identification by mass spectrometry. For each identified protein, the ratio of peptide spectral counts from the alk-16 and DMSO samples was plotted. Several known palmitoylated proteins are shown in black. IFITM3, the highest-ranked candidate palmitoylated protein, is shown in red.
Figure 2. IFITM3 is S-palmitoylated on membrane-proximal cysteine residues
(a) Ifitm3 mRnA expression in DC2.4 cells after 4 h IFn-α treatment as measured by qRt-PCR. NT, non-treated. (b) predicted topology of IFITM3 showing the location of cysteine residues (gray shading) with respect to transmembrane domains. (c,d) HeLa cells were transfected with 2 μg vector, pCMV-HA-IFITM3 wild-type (WT) or cysteine mutants in six-well plates overnight and labeled with 50 μM alk-16 for 2 h. Cell lysates were subjected to anti-HA immunoprecipitation, reacted with az-rho by CuAAC, separated by SDS-PAGE and visualized by fluorescence gel scanning. Comparable protein loading was confirmed by anti-HA western blotting. In c, half of the indicated immunoprecipitations were treated with 2.5% (w/v) neutral NH2OH before boiling and gel loading to hydrolyze protein–alk-16 thioester linkages.
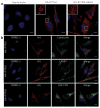
Figure 3. Palmitoylation-dependent clustering of IFITM3 in the ER
(a,b) HeLa cells grown on coverslips in 12-well plates were transfected with 1 μg pCMV-HA, pCMV-HA-IFITM3 or pCMV-HA-IFITM3-palmΔ and stained with anti-HA (red) and TOPRO-3 (blue). Insets are enlargements of the white-squared regions. Scale bars represent 10 μm. (b) Cells were also stained with antibodies against cellular markers, calreticulin and LAMP1 (green), or were co-transfected with GFP-CD9 (green).
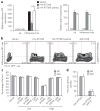
Figure 4. Antiviral activity of IFITM3 is regulated by palmitoylation
(a) HeLa cells grown in six-well plates were transfected with 2 μg pCMV-HA, pCMV-HA-IFITM3 or pCMV-HA-IFITM3-palmΔ and infected with influenza virus (PR8 strain) with multiplicity of infection (MOI) of 1 for 6 h. ND, not detected; NI, non-infected. Influenza virus NP mRNA levels and Ifitm3 expression levels were examined by qRT-PCR. *P = 0.027 by Student’s _t_-test; error represents s.d., n = 3. (b) HeLa cells grown in 12-well plates were transfected with 1 μg pCMV-HA, pCMV-HA-IFITM3, pCMV-HA-IFITM3-palmΔ or pEGFP-C1-CD9 and infected with influenza virus at an MOI of 1 for 6 h. Virus NP and IFITM3 protein levels were examined by flow cytometry using anti-NP and anti-HA staining, respectively. (c) HEK293T cells grown in 12-well plates were transfected with 1 μg pCMV-HA, pCMV-HA-IFITM3, pCMV-HA-IFITM3-palmΔ or pEGFP-C1-CD9 and infected with influenza virus at an MOI of 1 for 6 h. Non-transfected and transfected cells expressing the proteins of interest from the same culture were gated on (labeled “low” and “high,” respectively) and analyzed for the percentage of these cells that were infected (Supplementary Fig. 8). Data from flow cytometric analysis of cells staining positive for influenza NP or NS1. (d) percentages in c were normalized such that the difference in infection rates for vector control and HA-IFITM3 high was set at 100% antiviral activity. Error in c and d represents s.d., n = 4. Note: the HA-tag epitope is derived from an H3 influenza virus strain and is not present in the PR8 strain of H1N1 influenza virus.
Similar articles
- The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity.
McMichael TM, Zhang L, Chemudupati M, Hach JC, Kenney AD, Hang HC, Yount JS. McMichael TM, et al. J Biol Chem. 2017 Dec 29;292(52):21517-21526. doi: 10.1074/jbc.M117.800482. Epub 2017 Oct 27. J Biol Chem. 2017. PMID: 29079573 Free PMC article. - Discovery and Characterization of IFITM _S_-Palmitoylation.
Das T, Hang HC. Das T, et al. Viruses. 2023 Nov 28;15(12):2329. doi: 10.3390/v15122329. Viruses. 2023. PMID: 38140570 Free PMC article. Review. - Positive Regulation of the Antiviral Activity of Interferon-Induced Transmembrane Protein 3 by S-Palmitoylation.
Wen S, Song Y, Li C, Jin N, Zhai J, Lu H. Wen S, et al. Front Immunol. 2022 Jun 13;13:919477. doi: 10.3389/fimmu.2022.919477. eCollection 2022. Front Immunol. 2022. PMID: 35769480 Free PMC article. Review. - S-palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus.
Yount JS, Karssemeijer RA, Hang HC. Yount JS, et al. J Biol Chem. 2012 Jun 1;287(23):19631-41. doi: 10.1074/jbc.M112.362095. Epub 2012 Apr 17. J Biol Chem. 2012. PMID: 22511783 Free PMC article. - Site-Specific Lipidation Enhances IFITM3 Membrane Interactions and Antiviral Activity.
Garst EH, Lee H, Das T, Bhattacharya S, Percher A, Wiewiora R, Witte IP, Li Y, Peng T, Im W, Hang HC. Garst EH, et al. ACS Chem Biol. 2021 May 21;16(5):844-856. doi: 10.1021/acschembio.1c00013. Epub 2021 Apr 22. ACS Chem Biol. 2021. PMID: 33887136 Free PMC article.
Cited by
- Protein _S_-palmitoylation in immunity.
Das T, Yount JS, Hang HC. Das T, et al. Open Biol. 2021 Mar;11(3):200411. doi: 10.1098/rsob.200411. Epub 2021 Mar 3. Open Biol. 2021. PMID: 33653086 Free PMC article. Review. - The dispanins: a novel gene family of ancient origin that contains 14 human members.
Sällman Almén M, Bringeland N, Fredriksson R, Schiöth HB. Sällman Almén M, et al. PLoS One. 2012;7(2):e31961. doi: 10.1371/journal.pone.0031961. Epub 2012 Feb 20. PLoS One. 2012. PMID: 22363774 Free PMC article. - Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3.
Ling S, Zhang C, Wang W, Cai X, Yu L, Wu F, Zhang L, Tian C. Ling S, et al. Sci Rep. 2016 Apr 5;6:24029. doi: 10.1038/srep24029. Sci Rep. 2016. PMID: 27046158 Free PMC article. - Role of Post-translational Modifications in Influenza A Virus Life Cycle and Host Innate Immune Response.
Hu J, Zhang L, Liu X. Hu J, et al. Front Microbiol. 2020 Sep 4;11:517461. doi: 10.3389/fmicb.2020.517461. eCollection 2020. Front Microbiol. 2020. PMID: 33013775 Free PMC article. Review. - Characteristics of IFITM, the newly identified IFN-inducible anti-HIV-1 family proteins.
Chutiwitoonchai N, Hiyoshi M, Hiyoshi-Yoshidomi Y, Hashimoto M, Tokunaga K, Suzu S. Chutiwitoonchai N, et al. Microbes Infect. 2013 Apr;15(4):280-90. doi: 10.1016/j.micinf.2012.12.003. Epub 2013 Jan 30. Microbes Infect. 2013. PMID: 23376165 Free PMC article.
References
- Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. - PubMed
- Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7:355–366. - PubMed
- Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. - PubMed
- Charron G, Wilson J, Hang HC. Chemical tools for understanding protein lipidation in eukaryotes. Curr Opin Chem Biol. 2009;13:382–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI041111/AI/NIAID NIH HHS/United States
- AI041111/AI/NIAID NIH HHS/United States
- 1R21DA025751-01/DA/NIDA NIH HHS/United States
- R01 AI083284/AI/NIAID NIH HHS/United States
- R21 DA025751/DA/NIDA NIH HHS/United States
- AI083284-01/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials