Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy - PubMed (original) (raw)
doi: 10.1038/ng.619. Epub 2010 Jul 4.
Barry S Taylor, Shantanu Banerji, Alexis H Ramos, Mariana Lagos-Quintana, Penelope L Decarolis, Kinjal Shah, Nicholas D Socci, Barbara A Weir, Alan Ho, Derek Y Chiang, Boris Reva, Craig H Mermel, Gad Getz, Yevgenyi Antipin, Rameen Beroukhim, John E Major, Charles Hatton, Richard Nicoletti, Megan Hanna, Ted Sharpe, Tim J Fennell, Kristian Cibulskis, Robert C Onofrio, Tsuyoshi Saito, Neerav Shukla, Christopher Lau, Sven Nelander, Serena J Silver, Carrie Sougnez, Agnes Viale, Wendy Winckler, Robert G Maki, Levi A Garraway, Alex Lash, Heidi Greulich, David E Root, William R Sellers, Gary K Schwartz, Cristina R Antonescu, Eric S Lander, Harold E Varmus, Marc Ladanyi, Chris Sander, Matthew Meyerson, Samuel Singer
Affiliations
- PMID: 20601955
- PMCID: PMC2911503
- DOI: 10.1038/ng.619
Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy
Jordi Barretina et al. Nat Genet. 2010 Aug.
Abstract
Soft-tissue sarcomas, which result in approximately 10,700 diagnoses and 3,800 deaths per year in the United States, show remarkable histologic diversity, with more than 50 recognized subtypes. However, knowledge of their genomic alterations is limited. We describe an integrative analysis of DNA sequence, copy number and mRNA expression in 207 samples encompassing seven major subtypes. Frequently mutated genes included TP53 (17% of pleomorphic liposarcomas), NF1 (10.5% of myxofibrosarcomas and 8% of pleomorphic liposarcomas) and PIK3CA (18% of myxoid/round-cell liposarcomas, or MRCs). PIK3CA mutations in MRCs were associated with Akt activation and poor clinical outcomes. In myxofibrosarcomas and pleomorphic liposarcomas, we found both point mutations and genomic deletions affecting the tumor suppressor NF1. Finally, we found that short hairpin RNA (shRNA)-based knockdown of several genes amplified in dedifferentiated liposarcoma, including CDK4 and YEATS4, decreased cell proliferation. Our study yields a detailed map of molecular alterations across diverse sarcoma subtypes and suggests potential subtype-specific targets for therapy.
Figures
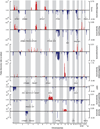
Figure 1. Nucleotide and copy number alterations in soft-tissue sarcoma subtypes
The statistical significance of genomic aberrations for each subtype is shown. RAE q-values [left axis; for visualization, q-values ≤ 0.05 are considered significant, corresponding false discovery rate (FDR) ≤ 5%] and scores (right axis) for gains and amplifications (red) and losses and deletions (blue) are plotted across the genome (chromosomes indicated at bottom). Genes harboring somatic nucleotide alterations in this study are indicated in each subtype in which they were discovered (Table 2).
Figure 2. NF1 alterations in karyotypically complex sarcomas
A. Somatic mutations in the NF1 protein in myxofibrosarcoma and pleomorphic liposarcoma (black triangles) and the position of the RasGAP and Cral domains (dark and light green respectively) are juxtaposed to known mutations in malignant peripheral nerve sheath tumors (MPNSTs; open triangles). B. Transcript expression according to copy number and sequence status in myxofibrosarcoma and pleomorphic liposarcoma compared to normal adipose tissue samples (black/red and green respectively, log2 expression from Affymetrix array profiling data; p-value=1.94×10−5, ANOVA; mutated tumors are indicated). One of the two R304* mutant tumors lacked expression data.
Figure 3. Different effect of helical and kinase domain PIK3CA mutations on PI3K pathway activation and survival in myxoid/round-cell liposarcoma
A. Survival for patients with tumors that harbor helical-domain mutations (red) versus kinase-domain mutations (grey), and wildtype PIK3CA (blue). The analysis includes the 65 patients for whom outcome information was available. Patients with mutations in either the helical or the kinase domain had a shorter disease–specific survival compared to those with wildtype PIK3CA (p-value = 0.0363, log-rank test). The difference in disease-specific survival between patients with helical-domain mutant tumors and those with wildtype PIK3CA tumors was significant (p-value=0.013, log-rank test). B. Western blots of myxoid/round-cell liposarcoma tumor lysates comparing the phosphorylation levels of Akt, PRAS40, and S6 kinase, as well as their protein levels, in patients with wild-type PIK3CA or with mutations in PIK3CA helical or kinase domains.
Figure 4. Genes whose knockdown is anti-proliferative in dedifferentiated liposarcoma and the consequences of CDK4, MDM2 and YEATS4 knockdown in dedifferentiated liposarcoma
(A) Integrated profile of statistically significant genomic gains/amplifications as assessed by both RAE and GISTIC (combined as described in Methods; FDR, false-discovery rate) is followed by a heatmap of copy number segmentation on 12q13.2-q32.1 in 50 patient samples of dedifferentiated liposarcomas (red is amplification, blue is deletion, each row indicates one tumor sample). Below is the position of genes from our screen encoded by this region of 12q whose knockdown is anti-proliferative in dedifferentiated liposarcoma. Bold gene symbols indicate those whose amplification produced over-expression of its transcript or those over-expressed in tumor relative to normal adipose tissue. Genes in green are highlighted in panels B–C and E. Alternative genomic regions encoding genes not on 12q whose knockdown is anti-proliferative are also included. (B) Effect of three validated shRNAs targeting CDK4 on the proliferation of two cell lines, LPS141 and DDLS8817, at various time points (x-axis) with negative controls (pLKO empty vector and GFP473). Below are western blots showing the effect of shRNAs on levels of CDK4 protein (as indicated). (C) G1 arrest induced in LPS141 and DDLS8817 cell lines by treatment with the CDK4/CDK6 inhibitor PD0332991. MDA-MB-435 (Rb-positive) and H2009 (Rb-negative) were included as sensitive and insensitive controls. Error bars are s.d. of replicate measurements. (D–E) As in panel (B), effect on proliferation of three shRNAs targeting MDM2 (panel D) and YEATS4 (panel E) (negative controls: pLKO empty vector and scrambled shRNA) where each targeting shRNA resulted in reduced protein levels (at bottom). Error bars are propagated error from the ratio of mean and s.d. of measurements/replicates to time 0.
Similar articles
- Gene expression analysis of soft tissue sarcomas: characterization and reclassification of malignant fibrous histiocytoma.
Nakayama R, Nemoto T, Takahashi H, Ohta T, Kawai A, Seki K, Yoshida T, Toyama Y, Ichikawa H, Hasegawa T. Nakayama R, et al. Mod Pathol. 2007 Jul;20(7):749-59. doi: 10.1038/modpathol.3800794. Epub 2007 Apr 27. Mod Pathol. 2007. PMID: 17464315 - Cytogenetic analysis of 46 pleomorphic soft tissue sarcomas and correlation with morphologic and clinical features: a report of the CHAMP Study Group. Chromosomes and MorPhology.
Mertens F, Fletcher CD, Dal Cin P, De Wever I, Mandahl N, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, Van den Berghe H, Vanni R, Willén H. Mertens F, et al. Genes Chromosomes Cancer. 1998 May;22(1):16-25. doi: 10.1002/(sici)1098-2264(199805)22:1<16::aid-gcc3>3.0.co;2-a. Genes Chromosomes Cancer. 1998. PMID: 9591630 - Analysis of germline and tumor mutations of p53 gene in familial occurrence of soft tissue sarcomas.
Kudawara I, Matsumine A, Ohzono K. Kudawara I, et al. J Surg Oncol. 2007 Mar 15;95(4):347-50. doi: 10.1002/jso.20720. J Surg Oncol. 2007. PMID: 17192950 - Lipomatous tumors.
Weiss SW. Weiss SW. Monogr Pathol. 1996;38:207-39. Monogr Pathol. 1996. PMID: 8744279 Review. - [Pleomorphic high-grade soft tissue sarcomas: is the subclassification up to date?].
Mechtersheimer G, Renner M, Penzel R, Schirmacher P. Mechtersheimer G, et al. Pathologe. 2011 Feb;32(1):47-56. doi: 10.1007/s00292-010-1400-4. Pathologe. 2011. PMID: 21234572 Review. German.
Cited by
- PTEN pathogenic variants are associated with poor prognosis in patients with advanced soft tissue sarcoma.
Pan M, Zhou MY, Jiang C, Zhang Z, Bui N, Bien J, Siy A, Achacoso N, Solorzano AV, Tse P, Chung E, Hu W, Thomas S, Ganjoo K, Habel LA. Pan M, et al. BJC Rep. 2024 Jan 30;2(1):9. doi: 10.1038/s44276-023-00029-3. BJC Rep. 2024. PMID: 39516677 - FUS::DDIT3 Fusion Protein in the Development of Myxoid Liposarcoma and Possible Implications for Therapy.
Hou X, Shi W, Luo W, Luo Y, Huang X, Li J, Ji N, Chen Q. Hou X, et al. Biomolecules. 2024 Oct 14;14(10):1297. doi: 10.3390/biom14101297. Biomolecules. 2024. PMID: 39456230 Free PMC article. Review. - Establishment and characterization of 18 Sarcoma Cell Lines: Unraveling the Molecular Mechanisms of Doxorubicin Resistance in Sarcoma Cell Lines.
Cho YE, Kim SC, Kim HJ, Han I, Ku JL. Cho YE, et al. J Transl Med. 2024 Oct 2;22(1):889. doi: 10.1186/s12967-024-05700-y. J Transl Med. 2024. PMID: 39358756 Free PMC article. - Atypical Spindle Cell/Pleomorphic Lipomatous Tumor: A Review and Update.
Nishio J, Nakayama S, Chijiiwa Y, Koga M, Aoki M. Nishio J, et al. Cancers (Basel). 2024 Sep 13;16(18):3146. doi: 10.3390/cancers16183146. Cancers (Basel). 2024. PMID: 39335118 Free PMC article. Review. - Novel MEN1-associated retroperitoneal pleomorphic liposarcoma.
McNicoll CF, Belmonte J, Nir I, Ferguson BD. McNicoll CF, et al. Rare Tumors. 2024 Sep 19;16:20363613241286934. doi: 10.1177/20363613241286934. eCollection 2024. Rare Tumors. 2024. PMID: 39314235 Free PMC article.
References
- Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. - PubMed
- Fletcher C, Unni K, Mertens F, editors. Pathology and Genetics of Tumors of Soft Tissue and Bone. Lyon: International Agency for Research on Cancer Press; 2002. p. 427.
- Heinrich MC, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. - PubMed
- Hirota S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. - PubMed
- Demetri GD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. - PubMed
Publication types
MeSH terms
Grants and funding
- U54 HG003067/HG/NHGRI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- K08 CA122833/CA/NCI NIH HHS/United States
- P01 CA047179-19/CA/NCI NIH HHS/United States
- P01 CA047179/CA/NCI NIH HHS/United States
- P50 CA140146/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous