Regulation of microRNA expression and abundance during lymphopoiesis - PubMed (original) (raw)
. 2010 Jun 25;32(6):828-39.
doi: 10.1016/j.immuni.2010.05.009. Epub 2010 Jun 3.
Wolfgang Resch, Arito Yamane, Nan Kuo, Zhiyu Li, Tirtha Chakraborty, Lai Wei, Arian Laurence, Tomoharu Yasuda, Siying Peng, Jane Hu-Li, Kristina Lu, Wendy Dubois, Yoshiaki Kitamura, Nicolas Charles, Hong-wei Sun, Stefan Muljo, Pamela L Schwartzberg, William E Paul, John O'Shea, Klaus Rajewsky, Rafael Casellas
Affiliations
- PMID: 20605486
- PMCID: PMC2909788
- DOI: 10.1016/j.immuni.2010.05.009
Regulation of microRNA expression and abundance during lymphopoiesis
Stefan Kuchen et al. Immunity. 2010.
Abstract
Although the cellular concentration of miRNAs is critical to their function, how miRNA expression and abundance are regulated during ontogeny is unclear. We applied miRNA-, mRNA-, and ChIP-Seq to characterize the microRNome during lymphopoiesis within the context of the transcriptome and epigenome. We show that lymphocyte-specific miRNAs are either tightly controlled by polycomb group-mediated H3K27me3 or maintained in a semi-activated epigenetic state prior to full expression. Because of miRNA biogenesis, the cellular concentration of mature miRNAs does not typically reflect transcriptional changes. However, we uncover a subset of miRNAs for which abundance is dictated by miRNA gene expression. We confirm that concentration of 5p and 3p miRNA strands depends largely on free energy properties of miRNA duplexes. Unexpectedly, we also find that miRNA strand accumulation can be developmentally regulated. Our data provide a comprehensive map of immunity's microRNome and reveal the underlying epigenetic and transcriptional forces that shape miRNA homeostasis.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
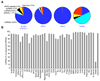
Figure 1. sRNA microsequencing of hematopoietic and non-hematopoietic cells
(A) Pie charts representing the distribution of the six different classes of sRNAs measured from LPS+IL-4 activated B cells, HPCs, heart, and testes. (B) Percentage of miRNAs of total sRNA sequences that had at least one perfect alignment to the mouse reference genome (mm9). Excluding testes, the miRNA mean for all cell types and tissues analyzed was 91.1%. Abbreviations: MEFs, mouse embryonic fibroblasts; S.Glands, salivary glands; ESCs, embryonic stem cells; HPCs, hematopoietic progenitor cells; proB, progenitor B220loCD43+ B cells; preB, B220loCD25+IgM− bone marrow B cells; B1, peritoneal B220loIgMhi B cells; MZ, IgMhiCR1CR2hi marginal zone B cells; GC, CD95hiB220+ germinal center B cells; PCs, CD138hiB220− plasma cells; LPS+IL-4, mature B220+CD43− splenic B cells activated ex-vivo for 72 hours in the presence of lipopolysaccharide and interleukin 4; LPS+α-δ-D, mature B cells activated ex-vivo for 72 hours in the presence of lipopolysaccharide and dextran conjugated anti-IgD; ConA, CD8+ T cells activated ex-vivo in the presence of concanavalin A for 72 hours; iTregs, ex-vivo induced regulatory T cells; nTregs, natural regulatory T cells isolated by cell sorting.
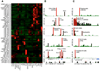
Figure 2. miRNA signatures of the mouse immune system, embryonic cells, and adult tissues
(A) Heat map showing expression of 375 miRNAs (detected at >100 TPM) in mouse hematopoietic and non-hematopoietic cells based on hierarchical clustering analysis. miRNAs enriched in a particular sample are depicted with red bars, while depleted miRNAs are depicted with green bars. B and T cell populations outlined in Table 1 were combined into a single group. (B) Percentage of let-7a (grey line), let-7c (red line), and let-7f (blue line). The total let-7 sequence tags were set to 100%. Let-7f and let-7c miRNA abundance is inversely proportional in hematopoietic cells (light blue box), whereas roughly equivalent in non-hematopoietic cells and tissues (light red box). (C) Expression profiles of miRNAs enriched in neutrophils. Two independently processed neutrophil samples (1 and 2) are shown and the highest TPM value of the two is given in parenthesis. (D) Examples of miRNAs predominantly expressed in HPCs, mast cells, and NK cells. For novel miRNAs, their genomic location is schematized. Orientation of miRNAs and genes is represented with red and black arrowheads respectively. Mammalian phylogenetic conservation is depicted with dense black bar graphs based on UCSC PhastCons30Way algorithm.
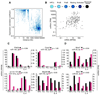
Figure 3. miRNA signatures of the mouse B and T cell compartments
(A) Heat map depicts expression of 49 lymphocyte-specific miRNAs. (B–C) Examples of representative miRNAs displaying specific expression in T or B lymphocytes. Only the highest TPM value is provided in each of the graphs.
Figure 4. Epigenetic control of miRNA expression in lymphocytes
(A) Chromatin modifications associated with promoter and gene body of Sgcz (encoding the heart-specific miR-383) during lymphopoiesis. Silencing is achieved via trimethylation of lysines 27 and 9 of histone H3, and lysine 20 of histone H4. Top bar graph provides expression profiles (in TPM) of miR-383_5p. (B) The ubiquitously expressed let-7g gene displays chromatin modifications associated with active genes, including RNA polII and CBP-p300 recruitment. Bar graph plots let-7g_5p. (C) Example of a hematopoietic miRNA gene, miR-155, induced at late stages of development but depleted of H3K27me3 in progenitor cells. Modification patterns of H3K14Ac, H3K36me3, H3K36Ac, H3K79me2, H2BK12Ac, and H3K27me3, as well as polII and p300 recruitment are shown for mature resting (black bars) and LPS+IL-4 activated (red bars) B cells. Values represent the total number of sequence tags aligned from 2.5Kb upstream of the gene transcription start site to its transcription termination site. Bar graph depicts expression profiles of miR-155_5p. (D) Example of a hematopoietic miRNA gene, miR-139, that retains H3K27me3 inhibitory mark up until full gene expression is invoked. Chromatin modifications H3K27me3 and H3K36me3 are shown and bar graph depicts miR-139_5p TPM values.
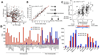
Figure 5. miRNA abundance in B cell development as a function of transcription
(A) Correlation between expression and chromosome distance separating mouse miRNAs. Data points represent all possible pair combinations for miRNAs located in the same chromosome. The y-axis provides the Pearson correlation coefficient calculated for tissue expression patterns, while the x-axis gives the physical distance between the miRNAs in kilobases (Kb). (B) The transcriptome of HPCs, proB cells, preB cells, mature resting, LPS+IL-4 activated, and germinal center B cells as determined by deep sequencing and normalized as RPKM values. Each data point represents one of 126 mRNA-miRNA pairs (Table S3) at any one of the six B cell developmental stages. (C) Examples of mRNA/miRNA pairs that show correlation (ρ > 0.7, upper row) or no correlation (ρ < 0.7, lower row) across the six B cell developmental stages. Black bars represent mRNAs RPKM values (left y-axis), while magenta bars depict miRNAs TPM values (right y-axis). (D) Examples of miRNAs with one strand (5p in both cases) showing greater correlation to mRNA expression profiles than the other.
Figure 6. 5p:3p fluctuations during lymphocyte development
(A) 5p vs. 3p species abundance of mouse miRNAs (graphed as log2 of TPM values). Closed red circles depict miRNAs entirely conserved between mouse and human. Dotted lines delimit all miRNAs that display roughly equal numbers of 5p and 3p species. (B) Correlation of 5p/3p ratios between mouse and human. Human miRNAs were microsequenced from 6 tonsillar B cell populations isolated by cell sorting: naïve, pre-germinal center, centrocytes, centroblasts, plasma cells, and memory B cells. For mouse, the entire panel of miRNA samples was used. Only miRNAs entirely conserved between the two species were plotted. Both the Spearman correlation and P values are provided. (C) Comparative analysis of miRNA 5p and 3p strand abundance (y-axis) vs. dissociation energy of 5’ and 3’ miRNA duplex ends (measured in Kcal/mol using RNAduplex software). The plot shows that as the relative thermodynamic stability of the miRNA duplex 3’ end increases (represented by positive values on the x-axis), its 5’ end is preferentially unwound, leading to asymmetric uploading of its 5p strand into the RISC complex and an overall increase in 5p cellular abundance (positive end of the y-axis). Data at the negative end of the x and y-axes provide the counterexample. (D) Left plot: relative abundance of miR-30c2_5p (red bars) and 3p (blue bars) strands across all cells and samples examined. Hematopoietic (light blue box) and non-hematopoietic (light red box) cells can be subdivided according to the absence or presence of miR-30c2_3p. Right plot: analysis of miR-150_5p (red bars) and 3p (blue bars) distribution in various B and T cell populations. Mature (2) and CD8 (2) refer to duplicate samples.
Similar articles
- Dynamic microRNA gene transcription and processing during T cell development.
Kirigin FF, Lindstedt K, Sellars M, Ciofani M, Low SL, Jones L, Bell F, Pauli F, Bonneau R, Myers RM, Littman DR, Chong MM. Kirigin FF, et al. J Immunol. 2012 Apr 1;188(7):3257-67. doi: 10.4049/jimmunol.1103175. Epub 2012 Feb 29. J Immunol. 2012. PMID: 22379031 Free PMC article. - Integration of microRNA signatures of distinct mammary epithelial cell types with their gene expression and epigenetic portraits.
Pal B, Chen Y, Bert A, Hu Y, Sheridan JM, Beck T, Shi W, Satterley K, Jamieson P, Goodall GJ, Lindeman GJ, Smyth GK, Visvader JE. Pal B, et al. Breast Cancer Res. 2015 Jun 18;17(1):85. doi: 10.1186/s13058-015-0585-0. Breast Cancer Res. 2015. PMID: 26080807 Free PMC article. - Novel expression profiles of microRNAs suggest that specific miRNAs regulate gene expression for the sexual maturation of female Schistosoma japonicum after pairing.
Sun J, Wang S, Li C, Ren Y, Wang J. Sun J, et al. Parasit Vectors. 2014 Apr 10;7:177. doi: 10.1186/1756-3305-7-177. Parasit Vectors. 2014. PMID: 24721600 Free PMC article. - Modulators of MicroRNA Function in the Immune System.
Jia Y, Wei Y. Jia Y, et al. Int J Mol Sci. 2020 Mar 29;21(7):2357. doi: 10.3390/ijms21072357. Int J Mol Sci. 2020. PMID: 32235299 Free PMC article. Review. - Epigenetic architecture and miRNA: reciprocal regulators.
Wiklund ED, Kjems J, Clark SJ. Wiklund ED, et al. Epigenomics. 2010 Dec;2(6):823-40. doi: 10.2217/epi.10.51. Epigenomics. 2010. PMID: 22122085 Review.
Cited by
- Modulation of Immunosuppression by Oligonucleotide-Based Molecules and Small Molecules Targeting Myeloid-Derived Suppressor Cells.
Lim J, Lee A, Lee HG, Lim JS. Lim J, et al. Biomol Ther (Seoul). 2020 Jan 1;28(1):1-17. doi: 10.4062/biomolther.2019.069. Biomol Ther (Seoul). 2020. PMID: 31431006 Free PMC article. Review. - Human AML activates the aryl hydrocarbon receptor pathway to impair NK cell development and function.
Scoville SD, Nalin AP, Chen L, Chen L, Zhang MH, McConnell K, Beceiro Casas S, Ernst G, Traboulsi AA, Hashi N, Williams M, Zhang X, Hughes T, Mishra A, Benson DM, Saultz JN, Yu J, Freud AG, Caligiuri MA, Mundy-Bosse BL. Scoville SD, et al. Blood. 2018 Oct 25;132(17):1792-1804. doi: 10.1182/blood-2018-03-838474. Epub 2018 Aug 29. Blood. 2018. PMID: 30158248 Free PMC article. - Identification of miR-128 Target mRNAs That Are Expressed in B Cells Using a Modified Dual Luciferase Vector.
Schreiber S, Daum P, Danzer H, Hauke M, Jäck HM, Wittmann J. Schreiber S, et al. Biomolecules. 2023 Oct 13;13(10):1517. doi: 10.3390/biom13101517. Biomolecules. 2023. PMID: 37892199 Free PMC article. - MiR-15b and miR-322 inhibit SETD3 expression to repress muscle cell differentiation.
Zhao MJ, Xie J, Shu WJ, Wang HY, Bi J, Jiang W, Du HN. Zhao MJ, et al. Cell Death Dis. 2019 Feb 22;10(3):183. doi: 10.1038/s41419-019-1432-5. Cell Death Dis. 2019. PMID: 30796205 Free PMC article. - MicroRNAs: the fine-tuners of Toll-like receptor signalling.
O'Neill LA, Sheedy FJ, McCoy CE. O'Neill LA, et al. Nat Rev Immunol. 2011 Mar;11(3):163-75. doi: 10.1038/nri2957. Epub 2011 Feb 18. Nat Rev Immunol. 2011. PMID: 21331081 Review.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases