Striatal microRNA controls cocaine intake through CREB signalling - PubMed (original) (raw)
Striatal microRNA controls cocaine intake through CREB signalling
Jonathan A Hollander et al. Nature. 2010.
Abstract
Cocaine addiction is characterized by a gradual loss of control over drug use, but the molecular mechanisms regulating vulnerability to this process remain unclear. Here we report that microRNA-212 (miR-212) is upregulated in the dorsal striatum of rats with a history of extended access to cocaine. Striatal miR-212 decreases responsiveness to the motivational properties of cocaine by markedly amplifying the stimulatory effects of the drug on cAMP response element binding protein (CREB) signalling. This action occurs through miR-212-enhanced Raf1 activity, resulting in adenylyl cyclase sensitization and increased expression of the essential CREB co-activator TORC (transducer of regulated CREB; also known as CRTC). Our findings indicate that striatal miR-212 signalling has a key role in determining vulnerability to cocaine addiction, reveal new molecular regulators that control the complex actions of cocaine in brain reward circuitries and provide an entirely new direction for the development of anti-addiction therapeutics based on the modulation of noncoding RNAs.
Figures
Figure 1. Increased striatal miR-212 expression in extended access rats
a, Expression analysis revealed that striatal miR-212 levels were increased in rats with extended access to cocaine self-administration. b, Taqman assay verified that striatal miR-212 levels were increased in extended access rats (_F_3,23=6.6, P<0.01; *P<0.05, **P<0.01, statistically significant compared with the extended access group). c, Relative amounts of p-CREB in the dorsal striatum were quantified by densitometry (_F_2,6=6.8, P<0.05; *P<0.05 compared with control). The lower panel is a representative immunoblot of the increased striatal p-CREB expression. Data are presented as mean ± SEM.
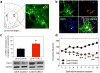
Figure 2. Dissociable effects of striatal miR-212 oncocaine intake
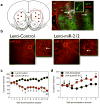
a, Red circles in left panel are locations at which viral infusions were targeted in dorsal striatum. The right panel is representative immunochemistry staining from a Lenti-miR-212 rat. Green is GFP from virus; red is the astrocyte marker glial fibrillary acidic protein (GFAP); Ctx, cortex; CC corpus callosum; LV, lateral ventricle; DSt, dorsal striatum; yellow arrows highlight the injector track used to deliver virus. Insert is an x80 confocal image of a virus-infected neuron. b, FISH was used to visualize striatal miR-212 expression (shown in red) in Lenti-Control and Lenti-miR-212 rats. c, Striatal miR-212 overexpression reverses the long-term trajectory of cocaine-taking behavior in rats with extended access (Virus x Session: _F_13,130=3.0, P<0.001; *P<0.05, **P<0.01 compared with intake on the same day in Lenti-Control rats). d, LNA-antimiR-212 delivered into dorsal striatum increases cocaine intake in extended access rats (LNA x Session: _F_4,36=5.3, P<0.005; *P<0.05, **P<0.01, compared with intake on the same day in the LNA-Scrambled rats). Data are presented as mean ± SEM.
Figure 3. miR-212 amplifies CREB signaling
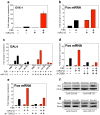
a, miR-212 potentiated CREB signaling engaged by forskolin (FSK),measured using an EVX-1 luciferase reporter (miRNA x FSK: _F_1,8=99.2, P<0.001). **b,** miR-212 potentiated forskolin-induced increases in Fos mRNA in HEK cells(miRNA x FSK: _F_1,8>1000, p<0.001). c, The effects of miR-212 on CREB signaling were abolished in GAL4-luciferase reporters containing phosphorylation-defective (ΔSer133) or TORC recruitment-defective (ΔR314) CREB mutants. d, A-CREB polypeptide abolished the effects of miR-212 on Fos mRNA expression. e, RNAi-mediated knockdown of TORC2 also abolished the effects of miR-212 on Fos mRNA expression. f, Protein levels of Fos were enhanced in dorsal striatum from Lenti-miR-212 rats with extended cocaine access compared with other groups. g, Striatal levels of Nurr1, another CREB-responsive gene, were also enhanced inLenti-miR-212 rats with extended access. Data are presented as mean ± SEM.
Figure 4. miR-212 stimulates core CREB signaling components
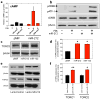
a, miR-212 potentiated cAMP accumulation in HEK cells (miRNA x FSK: F_2,8=4.8, P<0.05). b, miR-212 also increased p-CREB and the related p-ATF-1, without altering total-CREB. c,_ Representative immunoblots of the increased TORC2 levels in HEK cells. d, Relative amounts of TORC2 in HEK cells expressing miR-212, miR-132 or vector (pMIF) were quantified by densitometry (*P<0.05 compared with pMIF). e, Representative immunoblots showing that miR-212 increased striatal levels of TORC1 and TORC2. f, The relative amounts of striatal TORC1 and TORC2 were quantified by densitometry (*P<0.05, **P<0.01, compared with Lenti-Control).Data are presented as mean ± SEM.
Figure 5. miR-212 amplifies CREB signaling through Raf-1
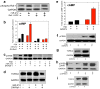
a, miR-212 activates Raf-1 signaling, reflected in increased levels of phosphorylated endogenous and exogenous (Raf-1-GFP) Raf-1 protein. b, Dominant-negative Raf-1 (DN-Raf-1) abolished the stimulatory effects of miR-212 on cAMP accumulation. c, DN-Raf-1abolished miR-212-induced increases in p-CREB. d, DN-Raf-1 also abolished miR-212-induced increases in TORC2. e, Enhancing Raf-1 signaling by pulsing cells with the Raf-1 inhibitor GW5074 potentiated cAMP accumulation. f, Potentiation of Raf-1 signaling increased p-CREB levels. g, Potentiation of Raf-1 signaling increased TORC2 levels. h, Knockdown of the Raf-1 repressor SPRED1, a target for miR-212, increased p-CREB levels. i, SPRED1 knockdown also increased TORC2 levels. Data are presented as mean ± SEM.
Figure 6. Striatal CREB:TORC signaling controls cocaine intake
a, Representative image of Lenti-TORC1-infected neurons (green) in the dorsal striatum. b, Representative high-magnification images of Lenti-TORC1 infected cells (green), demonstrating the viral-driven upregulation of TORC1 expression (red). Cell nuclei are shown in blue using DAPI staining. c, Relative amounts of TORC1in dorsal striatum were quantified by densitometry (*P<0.05). The lower panel is a representative immunoblot. d, Cocaine intake under extended access conditions is far lower in Lenti-TORC1 rats compared with Lenti-Controls(Virus x Session: _F_9,.90=2.9, P<0.001; *P<0.05, **P<0.001 compared with intake on the same day in Lenti-Control rats).Data are presented as mean ± SEM.
Comment in
- Neuroscience: MicroRNA knocks down cocaine.
Picciotto MR. Picciotto MR. Nature. 2010 Jul 8;466(7303):194-5. doi: 10.1038/466194a. Nature. 2010. PMID: 20613832 No abstract available.
Similar articles
- Neuroscience: MicroRNA knocks down cocaine.
Picciotto MR. Picciotto MR. Nature. 2010 Jul 8;466(7303):194-5. doi: 10.1038/466194a. Nature. 2010. PMID: 20613832 No abstract available. - Reflections on: "A general role for adaptations in G-Proteins and the cyclic AMP system in mediating the chronic actions of morphine and cocaine on neuronal function".
Nestler EJ. Nestler EJ. Brain Res. 2016 Aug 15;1645:71-4. doi: 10.1016/j.brainres.2015.12.039. Epub 2015 Dec 29. Brain Res. 2016. PMID: 26740398 Free PMC article. Review. - Antisense-induced reduction in nucleus accumbens cyclic AMP response element binding protein attenuates cocaine reinforcement.
Choi KH, Whisler K, Graham DL, Self DW. Choi KH, et al. Neuroscience. 2006;137(2):373-83. doi: 10.1016/j.neuroscience.2005.10.049. Epub 2005 Dec 15. Neuroscience. 2006. PMID: 16359811 - A role for calmodulin-stimulated adenylyl cyclases in cocaine sensitization.
DiRocco DP, Scheiner ZS, Sindreu CB, Chan GC, Storm DR. DiRocco DP, et al. J Neurosci. 2009 Feb 25;29(8):2393-403. doi: 10.1523/JNEUROSCI.4356-08.2009. J Neurosci. 2009. PMID: 19244515 Free PMC article. - Molecular, cellular, and structural mechanisms of cocaine addiction: a key role for microRNAs.
Jonkman S, Kenny PJ. Jonkman S, et al. Neuropsychopharmacology. 2013 Jan;38(1):198-211. doi: 10.1038/npp.2012.120. Epub 2012 Sep 12. Neuropsychopharmacology. 2013. PMID: 22968819 Free PMC article. Review.
Cited by
- MicroRNAs shape the neuronal landscape.
McNeill E, Van Vactor D. McNeill E, et al. Neuron. 2012 Aug 9;75(3):363-79. doi: 10.1016/j.neuron.2012.07.005. Neuron. 2012. PMID: 22884321 Free PMC article. Review. - miR-132 and miR-942 Expression Levels in Children with Attention Deficit and Hyperactivity Disorder: A Controlled Study.
Coskun S, Karadag M, Gokcen C, Oztuzcu S. Coskun S, et al. Clin Psychopharmacol Neurosci. 2021 May 31;19(2):262-268. doi: 10.9758/cpn.2021.19.2.262. Clin Psychopharmacol Neurosci. 2021. PMID: 33888655 Free PMC article. - Improving translation of animal models of addiction and relapse by reverse translation.
Venniro M, Banks ML, Heilig M, Epstein DH, Shaham Y. Venniro M, et al. Nat Rev Neurosci. 2020 Nov;21(11):625-643. doi: 10.1038/s41583-020-0378-z. Epub 2020 Oct 6. Nat Rev Neurosci. 2020. PMID: 33024318 Review. - Comprehensive Analysis of Differential m6A RNA Methylomes in the Hippocampus of Cocaine-Conditioned Mice.
Xue A, Huang Y, Li M, Wei Q, Bu Q. Xue A, et al. Mol Neurobiol. 2021 Aug;58(8):3759-3768. doi: 10.1007/s12035-021-02363-4. Epub 2021 Apr 7. Mol Neurobiol. 2021. PMID: 33826069 - Histone demethylase retinoblastoma binding protein 2 is overexpressed in hepatocellular carcinoma and negatively regulated by hsa-miR-212.
Liang X, Zeng J, Wang L, Fang M, Wang Q, Zhao M, Xu X, Liu Z, Li W, Liu S, Yu H, Jia J, Chen C. Liang X, et al. PLoS One. 2013 Jul 29;8(7):e69784. doi: 10.1371/journal.pone.0069784. Print 2013. PLoS One. 2013. PMID: 23922798 Free PMC article.
References
- McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. - PubMed
- Kalivas PW, O’Brien CP. Drug addiction as a pathology of staged neuroplasticity. Neuropsychopharmacology. 2008;33:166–180. - PubMed
- Carlezon WAJ, et al. Regulation of cocaine reward by CREB. Science. 1998;282:2272–2275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 CA134121/CA/NCI NIH HHS/United States
- F32 DA024932/DA/NIDA NIH HHS/United States
- R01 DA025983/DA/NIDA NIH HHS/United States
- R01 DA025983-03/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous