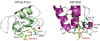Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b - PubMed (original) (raw)
Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b
Lei Zeng et al. Nature. 2010.
Abstract
Histone lysine acetylation and methylation have an important role during gene transcription in a chromatin context. Knowledge concerning the types of protein modules that can interact with acetyl-lysine has so far been limited to bromodomains. Recently, a tandem plant homeodomain (PHD) finger (PHD1-PHD2, or PHD12) of human DPF3b, which functions in association with the BAF chromatin remodelling complex to initiate gene transcription during heart and muscle development, was reported to bind histones H3 and H4 in an acetylation-sensitive manner, making it the first alternative to bromodomains for acetyl-lysine binding. Here we report the structural mechanism of acetylated histone binding by the double PHD fingers of DPF3b. Our three-dimensional solution structures and biochemical analysis of DPF3b highlight the molecular basis of the integrated tandem PHD finger, which acts as one functional unit in the sequence-specific recognition of lysine-14-acetylated histone H3 (H3K14ac). Whereas the interaction with H3 is promoted by acetylation at lysine 14, it is inhibited by methylation at lysine 4, and these opposing influences are important during transcriptional activation of the mouse DPF3b target genes Pitx2 and Jmjd1c. Binding of this tandem protein module to chromatin can thus be regulated by different histone modifications during the initiation of gene transcription.
Figures
Figure 1. 3D structure of the tandem PHD finger of human DPF3b
(a) Domain organization of human DPF3 proteins, including 2/3 domain (light brown), nuclear localization signal (NLS, red), nuclear receptor interaction domain (NID, pink), C2H2 Krüppel-like zinc finger (purple), PHD finger 1 (green) and PHD finger 2 (blue). (b) 2D 1H-15N-HSQC NMR spectra showing DPF3b PHD12 binding to histone peptide H3K14ac (residues 1–18) or H4-NAc (residues 1–22), as illustrated by change of protein resonances between the free (black) and peptide bound (red) states. (c) Isothermal titration calorimetry measurements of PHD12 binding to different H3 and H4 peptides. (d) Ribbon diagram depicting the average minimized NMR structure of the PHD12 bound to an H3K14ac peptide (yellow) in front and top views. The zinc atoms are highlighted as red spheres. (e) Space filled PHD12 structure, highlighting the H3K14ac peptide (yellow) bound across the unified structure of PHD1 (green) and PHD2 (blue).
Figure 2. Structural mechanism of acetyl-lysine recognition by human DPF3b PHD12
(a) DPF3b PHD1 recognition of H3K14ac, as illustrated in the PHD12/H3K14ac complex structure with stereo ribbon view (left) and electrostatic potential surface of the protein (right). (b) DPF3b PHD2 recognition of the N-terminal residues of the H3K14ac peptide, depicted in stereo ribbon structure (left) and protein electrostatic potential surface representation (right). The zinc atoms are highlighted as red spheres.
Figure 3. Distinct modes of acetyl-lysine and H3 recognition by DPF3b PHD12 and other protein modules
Comparison of the structural features of acetyl-lysine binding pockets between DPF3b PHD12/H3K14ac (left) and CBP bromodomain/H4K20ac (right) complexes. The protein and peptide residues are color-coded in the same scheme as described in Fig. 2.
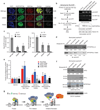
Figure 4. Acetylation and methylation modulated histone H3 binding by DPF3b PHD12 is important for gene transcriptional activation
(a) Co-localization of EGFP-DPF3b PHD12, or its acetyl-lysine binding deficient mutants, and H3K14ac in the C2C12 cell nucleus visualized by confocal fluorescence imaging. Calculated mean normalized total signal intensity value for each DPF3b is listed on the right (Supplementary Table 4). (b) Sequential chromatin immunoprecipitation (re-ChIP) assay (scheme, left) used to assess EGFP-DPF3b or its PHD12 mutants in interactions with H3K14ac at the mouse Pitx2 promoter site (right). (c) Quantitation of DNA recovered for Pitx2 and Jmjd1c promoters by qPCR following enrichment by immunoprecipitations with anti-sera against H3K14ac and seq-ChIP of EGFP/DPF3b with anti-EGFP agarose. Two non-DPF3b target genes Enpep in chromosome 3 and Arid5b in chromosome 10 were used in qChIP as negative controls to Pitx2 and Jmjd1c, respectively. (d) Assessing wild type DPF3b and mutant F264A binding to C-terminal biotinylated histone H3 peptides of different modifications in a peptide pull-down assay. Immunoblots depict H3 peptides binding to DPF3b from nuclear extracts of C2C12 cells transfected with the corresponding GFP/DPF3b plasmid. Signals in the immunoblots were quantified using NIH’s ImageJ software, and normalized to the input (1%) as 1.00. (e) Relative transcript levels of endogenous Pitx2 and Jmjd1c in C2C12 cells 2 days following transfections with wild type and mutant DPF3b cDNAs as indicated. (f) Evaluation of endogenous DPF2, HP1α and BPTF/FALZ in nuclear extracts of C2C12 cells binding to histone H3 peptides of different modifications. A single filter was used to determine retention of specific endogenous protein for each of the H3 peptides as shown. Separately, 5% of overall input for the peptide pull-down was monitored with β-actin. Blots were scanned and densitometry measurements were performed as described in d. (g) Schematic diagram illustrating modulation of human DPF3b PHD12 binding to histone H3 by site-specific lysine acetylation and methylation during poised, pre-initiation and initiation/activation stages of gene transcription. PHD1 and PHD2 of DPF3b are color-coded in yellow and blue, respectively. TF stands for a gene specific transcription factor, and TMC (orange) represents the transcriptional machinery complex.
Similar articles
- Crystal structure of DPF3b in complex with an acetylated histone peptide.
Li W, Zhao A, Tempel W, Loppnau P, Liu Y. Li W, et al. J Struct Biol. 2016 Sep;195(3):365-372. doi: 10.1016/j.jsb.2016.07.001. Epub 2016 Jul 16. J Struct Biol. 2016. PMID: 27402533 - Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14.
Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. Kasten M, et al. EMBO J. 2004 Mar 24;23(6):1348-59. doi: 10.1038/sj.emboj.7600143. Epub 2004 Mar 4. EMBO J. 2004. PMID: 15014446 Free PMC article. - Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF.
Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Li H, et al. Nature. 2006 Jul 6;442(7098):91-5. doi: 10.1038/nature04802. Epub 2006 May 21. Nature. 2006. PMID: 16728978 Free PMC article. - The PHD finger: a versatile epigenome reader.
Sanchez R, Zhou MM. Sanchez R, et al. Trends Biochem Sci. 2011 Jul;36(7):364-72. doi: 10.1016/j.tibs.2011.03.005. Epub 2011 Apr 21. Trends Biochem Sci. 2011. PMID: 21514168 Free PMC article. Review. - Atypical histone targets of PHD fingers.
Black JC, Kutateladze TG. Black JC, et al. J Biol Chem. 2023 Apr;299(4):104601. doi: 10.1016/j.jbc.2023.104601. Epub 2023 Mar 11. J Biol Chem. 2023. PMID: 36907441 Free PMC article. Review.
Cited by
- High-throughput strategy to identify inhibitors of histone-binding domains.
Wagner EK, Albaugh BN, Denu JM. Wagner EK, et al. Methods Enzymol. 2012;512:161-85. doi: 10.1016/B978-0-12-391940-3.00008-1. Methods Enzymol. 2012. PMID: 22910207 Free PMC article. - Comprehensive characterization of three classes of Arabidopsis SWI/SNF chromatin remodelling complexes.
Guo J, Cai G, Li YQ, Zhang YX, Su YN, Yuan DY, Zhang ZC, Liu ZZ, Cai XW, Guo J, Li L, Chen S, He XJ. Guo J, et al. Nat Plants. 2022 Dec;8(12):1423-1439. doi: 10.1038/s41477-022-01282-z. Epub 2022 Dec 5. Nat Plants. 2022. PMID: 36471048 - Sophisticated Conversations between Chromatin and Chromatin Remodelers, and Dissonances in Cancer.
Clapier CR. Clapier CR. Int J Mol Sci. 2021 May 25;22(11):5578. doi: 10.3390/ijms22115578. Int J Mol Sci. 2021. PMID: 34070411 Free PMC article. Review. - Chromatin Remodelers: From Function to Dysfunction.
Längst G, Manelyte L. Längst G, et al. Genes (Basel). 2015 Jun 12;6(2):299-324. doi: 10.3390/genes6020299. Genes (Basel). 2015. PMID: 26075616 Free PMC article. Review. - Family-Wide Characterization of Histone Binding Abilities of PHD Domains of AL Proteins in Arabidopsis thaliana.
Liang X, Lei M, Li F, Yang X, Zhou M, Li B, Cao Y, Gong S, Liu K, Liu J, Qi C, Liu Y. Liang X, et al. Protein J. 2018 Dec;37(6):531-538. doi: 10.1007/s10930-018-9796-4. Protein J. 2018. PMID: 30259302
References
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. - PubMed
- Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases