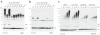Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne - PubMed (original) (raw)
Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne
Anja Bremm et al. Nat Struct Mol Biol. 2010 Aug.
Abstract
Ubiquitin is a versatile cellular signaling molecule that can form polymers of eight different linkages, and individual linkage types have been associated with distinct cellular functions. Though little is currently known about Lys11-linked ubiquitin chains, recent data indicate that they may be as abundant as Lys48 linkages and may be involved in vital cellular processes. Here we report the generation of Lys11-linked polyubiquitin in vitro, for which the Lys11-specific E2 enzyme UBE2S was fused to a ubiquitin binding domain. Crystallographic and NMR analyses of Lys11-linked diubiquitin reveal that Lys11-linked chains adopt compact conformations in which Ile44 is solvent exposed. Furthermore, we identify the OTU family deubiquitinase Cezanne as the first deubiquitinase with Lys11-linkage preference. Our data highlight the intrinsic specificity of the ubiquitin system that extends to Lys11-linked chains and emphasize that differentially linked polyubiquitin chains must be regarded as independent post-translational modifications.
Figures
Figure 1
UBE2S is a Lys11-specific E2 enzyme (a) UBE2S and (b) UBE2C were analyzed in autoubiquitination assays in the presence of E1, ubiquitin and Mg•ATP. The panel of single-Lys ubiquitin mutants reveals the intrinsic linkage specificity. Autoubiquitination is visualized with a polyclonal anti-ubiquitin antibody. UBE2S, but not UBE2C, autoubiquitinates and also assembles unattached Lys11-linked ubiquitin chains. (c) Time course assay for autoubiquitination by UBE2S. The reaction for wild-type (wt) and Lys11-only ubiquitin leads to similar high-molecular weight conjugates, while for the Lys-less (K0) and Lys63-only ubiquitin an equivalent pattern of multi-monoubiquitination is observed.
Figure 2
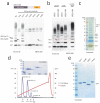
Assembly of Lys11-linked diubiquitin (a) Domain structure of UBE2S, and autoubiquitination reactions with UBE2S wild-type and catalytic mutants. (b) UBE2S autoubiquitination occurs in cis. Wild-type UBE2S was mixed with GST-tagged inactive UBE2SC95A, and after precipitation of the GST-tagged protein, ubiquitination in supernatant (left) and precipitate (right) is analyzed. (c) Removal of the Lys-rich tail of UBE2S decreases autoubiquitination while preserving Lys11 specificity. (d) Purification of Lys11-linked diubiquitin by cation exchange chromatography. The integrated peak area (mAU*ml) is indicated. A gel showing protein-containing fractions is shown as an inset.
Figure 3
Assembly of Lys11-linked tetraubiquitin (a) UBE2S engineering to increase yields of free Lys11-linked ubiquitin chains. The C-terminal tail was replaced with the ZnF-UBP domain of USP5/IsoT. The fusion protein assembles free chains of up to five ubiquitin molecules, yet it is less specific and also incorporates Lys63-linkages with wild-type and Lys63-only ubiquitin (indicated by arrows). (b) Incorporation of Lys63-linkages can be counteracted by using a K63R ubiquitin mutant, or by including the Lys63-specific DUB AMSH in the reaction, as observed by disappearance of the faster migrating Lys63-linkage contamination. (c) 5 μl aliquot of a 1 ml chain assembly reaction using 25 mg ubiquitin shows that di-, tri- and tetraubiquitin is generated in milligram quantities. (d) Cation exchange chromatography was used to purify Lys11-linked ubiquitin chains. The integrated peak area (mAU*ml) is specified. A gel showing protein-containing fractions is shown as an inset. (e) Purified ubiquitin tetramers of Lys11, Lys48, Lys63 and linear linkages have different electrophoretic mobility on 4-12% SDS-PAGE gels.
Figure 4
NMR Solution studies of Lys11-linked diubiquitin. (a) Overlay of 15N,1H HSQC spectra of ubiquitin K63R (red) onto Lys11-linked diubiquitin K63R (blue). The expansion illustrates the doubling of peaks observed for Lys29, Ile30, Asp32 and Lys33. The signal for Asp52 is unperturbed. (b, c) Weighted chemical shift perturbation according to residue number for Lys11-linked diubiquitin with both molecules 13C, 15N–labeled (blue, K63R ubiquitin mutant) or only labeled distally (orange, K11R ubiquitin mutant). Shown are chemical shift perturbations observed for doubled peaks calculated as the weighted difference between the chemical shift position in the Lys11-linked diubiquitin mutants and their respective monoubiquitin counterparts at pH7.2 (b) and pH 3.5 (c). Stars (*) indicate exchange broadened residues, and arrows indicate K29 and K33. (d) Combined chemical shift perturbation differences for Lys48- and Lys63-linked diubiquitin. This data was kindly provided by Takeshi Tenno .
Figure 5
Crystal structure of Lys11-linked diubiquitin (a) The crystal structure of Lys11-linked diubiquitin in two orientations. The proximal (orange) and distal (yellow) molecules interact through the ubiquitin helix, and the isopeptide linkage (shown in ball-and-stick representation, with red oxygen and blue nitrogen atoms) is at the surface of the dimer. (b) A semi-transparent surface colored blue for residues Ile44, Leu8 and Val70 shows that the hydrophobic patch is not involved in the interface. (c) Residues at the interface are shown in stick representation, and polar interactions of <3.5Å are shown with dotted lines. Water molecules are shown as purple spheres.
Figure 6
Hydrolysis of Lys11 linkages by DUBs. (a, b) Analysis of a panel of DUBs from all human DUB families against Lys11- (a) and Lys63-linked (b) diubiquitin. Two lanes per DUB in a silver stained gel indicate hydrolysis of ubiquitin dimers after 10 and 60 min, respectively. The Lys63-linked dimers were not tested against UCH family members as these were previously found to be inactive against this chain type . (c) The panel of DUBs was analyzed against Lys11-polyubiquitinated UBE2S and ubiquitin was visualized by Western blotting.
Figure 7
Cezanne cleaves Lys11-linkages preferentially (a) Phylogenetic tree of the OTU family DUBs , showing the close relationship of A20, TRABID and Cezanne. (b) Schematic presentation of domain structures of the OTU family DUBs Cezanne, A20 and TRABID. (c-e) Specificity analysis of Cezanne (c), A20 (d) and TRABID (e) against Lys11-, Lys48-, Lys63- and linear ubiquitin tetramers, resolved on 4-12% SDS-PAGE gels and visualized by silver staining. In this comparison, Cezanne shows specificity for Lys11-linked chains, while A20 and TRABID remain Lys48- and Lys63-specific, respectively. (f) Cezanne prefers Lys11-linkages in a mixture of differently linked ubiquitin tetramers. The different electrophoretic mobility of the individual tetraubiquitin molecules allows identification of the linkage type.
Similar articles
- Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Mevissen TET, Kulathu Y, Mulder MPC, Geurink PP, Maslen SL, Gersch M, Elliott PR, Burke JE, van Tol BDM, Akutsu M, Oualid FE, Kawasaki M, Freund SMV, Ovaa H, Komander D. Mevissen TET, et al. Nature. 2016 Oct 20;538(7625):402-405. doi: 10.1038/nature19836. Epub 2016 Oct 12. Nature. 2016. PMID: 27732584 Free PMC article. - Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling.
Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Varadan R, et al. J Biol Chem. 2004 Feb 20;279(8):7055-63. doi: 10.1074/jbc.M309184200. Epub 2003 Nov 25. J Biol Chem. 2004. PMID: 14645257 - Emerging roles for Lys11-linked polyubiquitin in cellular regulation.
Bremm A, Komander D. Bremm A, et al. Trends Biochem Sci. 2011 Jul;36(7):355-63. doi: 10.1016/j.tibs.2011.04.004. Epub 2011 Jun 7. Trends Biochem Sci. 2011. PMID: 21641804 Review. - Crosstalk between Lys63- and Lys11-polyubiquitin signaling at DNA damage sites is driven by Cezanne.
Wu X, Liu S, Sagum C, Chen J, Singh R, Chaturvedi A, Horton JR, Kashyap TR, Fushman D, Cheng X, Bedford MT, Wang B. Wu X, et al. Genes Dev. 2019 Dec 1;33(23-24):1702-1717. doi: 10.1101/gad.332395.119. Epub 2019 Nov 7. Genes Dev. 2019. PMID: 31699778 Free PMC article. - Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages.
Kulathu Y, Komander D. Kulathu Y, et al. Nat Rev Mol Cell Biol. 2012 Jul 23;13(8):508-23. doi: 10.1038/nrm3394. Nat Rev Mol Cell Biol. 2012. PMID: 22820888 Review.
Cited by
- Deciphering tissue-specific ubiquitylation by mass spectrometry.
Mayor U, Peng J. Mayor U, et al. Methods Mol Biol. 2012;832:65-80. doi: 10.1007/978-1-61779-474-2_3. Methods Mol Biol. 2012. PMID: 22350876 Free PMC article. - Uncovering DUB Selectivity through an Ion Mobility-Based Assessment of Ubiquitin Chain Isomers.
Shestoperova EI, Strieter ER. Shestoperova EI, et al. Anal Chem. 2023 Nov 28;95(47):17416-17423. doi: 10.1021/acs.analchem.3c04622. Epub 2023 Nov 14. Anal Chem. 2023. PMID: 37962301 Free PMC article. - The acetylase activity of Cdu1 regulates bacterial exit from infected cells by protecting Chlamydia effectors from degradation.
Bastidas RJ, Kędzior M, Davidson RK, Walsh SC, Dolat L, Sixt BS, Pruneda JN, Coers J, Valdivia RH. Bastidas RJ, et al. Elife. 2024 Feb 15;12:RP87386. doi: 10.7554/eLife.87386. Elife. 2024. PMID: 38358795 Free PMC article. - Preserving genome integrity: The vital role of SUMO-targeted ubiquitin ligases.
Han J, Mu Y, Huang J. Han J, et al. Cell Insight. 2023 Oct 23;2(6):100128. doi: 10.1016/j.cellin.2023.100128. eCollection 2023 Dec. Cell Insight. 2023. PMID: 38047137 Free PMC article. Review. - Oxygen-dependent asparagine hydroxylation of the ubiquitin-associated (UBA) domain in Cezanne regulates ubiquitin binding.
Mader J, Huber J, Bonn F, Dötsch V, Rogov VV, Bremm A. Mader J, et al. J Biol Chem. 2020 Feb 21;295(8):2160-2174. doi: 10.1074/jbc.RA119.010315. Epub 2020 Jan 14. J Biol Chem. 2020. PMID: 31937588 Free PMC article.
References
Experimental Procedures References
- Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. - PubMed
- Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–8. - PubMed
- Hajduk PJ, et al. NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J Med Chem. 1997;40:3144–50. - PubMed
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. - PubMed
References
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–53. - PubMed
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. - PubMed
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. - PubMed
- Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials