Structural basis for agonism and antagonism of hepatocyte growth factor - PubMed (original) (raw)
Structural basis for agonism and antagonism of hepatocyte growth factor
W David Tolbert et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Hepatocyte growth factor (HGF) is an activating ligand of the Met receptor tyrosine kinase, whose activity is essential for normal tissue development and organ regeneration but abnormal activation of Met has been implicated in growth, invasion, and metastasis of many types of solid tumors. HGF has two natural splice variants, NK1 and NK2, which contain the N-terminal domain (N) and the first kringle (K1) or the first two kringle domains of HGF. NK1, which is a Met agonist, forms a head-to-tail dimer complex in crystal structures and mutations in the NK1 dimer interface convert NK1 to a Met antagonist. In contrast, NK2 is a Met antagonist, capable of inhibiting HGF's activity in cell proliferation without clear mechanism. Here we report the crystal structure of NK2, which forms a "closed" monomeric conformation through interdomain interactions between the N- domain and the second kringle domain (K2). Mutations that were designed to open up the NK2 closed conformation by disrupting the N/K2 interface convert NK2 from a Met antagonist to an agonist. Remarkably, this mutated NK2 agonist can be converted back to an antagonist by a mutation that disrupts the NK1/NK1 dimer interface. These results reveal the molecular determinants that regulate the agonist/antagonist properties of HGF NK2 and provide critical insights into the dimerization mechanism that regulates the Met receptor activation by HGF.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
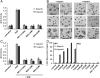
Fig. 1.
Heparin independent binding of NK2 to Met (A and B). Binding of NK1 and NK2 to Met, as determined by competition of the binding of biotinylated NK1 to Met in AlphaScreen Assays, in the absence (A) and presence (B) of heparin. Both NK1 and NK2 contain the K132E and R134E mutations that were designated as 1K1 (18). (C and D). Heparin independent binding of NK1 mutants to Met, as determined by AlphaScreen competition curves, for the Met567-NK1 interaction in the absence (A) and presence (B) of heparin. The NK1 mutants tested are: NK1(R134G) (black), NK1(A)—NK1(R134G,R73E) (red), NK1(B)—NK1(R134G,K58A,K60A,T61A,K62A) (blue), NK1(C)—NK1(R134G,K58E,K60E,T61A,K62E) (green), and NK1(D)—NK1(R134G,K58E,K60E,T61A,K62E,R73E) (orange). IC50 values derives from these curves are summarized in Table 1.
Fig. 2.
NK2 is a Met antagonist (A) uPA induction of MDCK cells by HGF (60 ng/mL), the NK1(Y124A) antagonist, and NK2. (B) MDCK cell scatter assays for HGF (60 ng/mL), NK1(Y124A) (1 μM), NK2 (0.0125 μM), or HGF + NK1 (Y124A), and HGF + NK2, in the presence or absence of heparin. (C) Inhibition of HGF-mediated uPA activation by NK2 and the NK1(Y124A) antagonist. Experiments are identical to (A) except that 60 ng/mL HGF was added to wells that were pretreated with NK1(Y124A) or NK2. (D) NK2 fails to induce Met dimerization regardless of the presence of heparin. NK1 and NK1 (Y124A) are used as positive and negative controls for their ability to induce Met-dimerization, which is assayed based on a modified AlphaScreen reported previously (8). Values are normalized to the dimerization signal of NK1 at 0.50 μM in the presence of heparin.
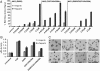
Fig. 3.
Crystal structure of NK2 (A) NK2 is a monomer. The overall structure of NK2 is represented by a ribbon diagram. Individual domains are designated by N for the N-terminal domain, K1 for the first kringle domain, and K2 for the second kringle domain. (B) The N-domain has a rigid structure. An overlay of the mouse (purple) and human (blue) N-domain crystal structures with the N-domain from the NK2 structure (green) shows the rigid fold of the N-domain. K1 and K2 domains of NK2 are omitted for clarity. (C) Kringle domain has a conserved rigid structure as shown by an overlay of K1 (purple) and K2 (green) domains of NK2. (D) The dimeric structure of NK1 (PDB ID 2QJ2) (blue) is presented for direct comparison of the monomeric NK2 structure (green) represented in (A). (E) The N/K2 interface. The interactions between intramolecular N and K2 domains are are shown as dashed yellow lines and disulfide bonds are shown as solid yellow lines. Nitrogen and oxygen atoms in side chain or main chain atoms are colored blue or red respectively. Only the alpha carbon residues are shown for the residues not involved in either interface. The first kringle domain of the N/K2 interface is omitted for clarity. All structure figures were made with Bobscript (–35) and Raster3D (36).
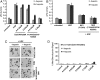
Fig. 4.
Conversion of NK2 to a Met agonist by disrupting the N/K2 interface (A) Mutations that disrupt the N/K2 interface promote the ability of NK2 to induce Met dimerization, which is assayed based on a modified AlphaScreen reported previously (8). Three different mutated NK2 are shown in the presence and absence of heparin. (B) Induction of uPA by HGF (60 ng/mL) and the three NK2 mutated variants (0.0125 μM) in the presence and absence of heparin in MDCK cells. The D257A/N258A NK2 has nearly the same ability to induce uPA activation as HGF. (C) Scattering of MDCK cells by the D257A/N258A NK2 mutant (0.0125 μM) and HGF (60 ng/mL). Top row (-heparin), bottom row (+heparin).
Fig. 5.
Conversion of the D257A/N258A NK2 from a Met agonist to a Met antagonist by mutating Y124A in the NK1 interface. (A) uPA induction assays in MDCK cells for HGF (60 ng/mL), the D257A/N258A NK2, and the D257A/N258A NK2 containing the Y124A mutation in the NK1 interface show the Y124A mutation abolishes the ability of the D257A/N258A NK2 to activate uPA. (B) An antagonist assay shows the ability of Y124A mutated NK1 and NK2 (D257A/N258A) to inhibit HGF-mediated activation of uPA in MDCK cells. (C) An antagonist assay shows the ability of Y124A mutated NK1 and NK2 (D257A/N258A) to inhibit HGF-mediated scattering of MDCK cells. The Y124A NK1 and NK2 were used as antagonist controls. (D) The Y124A mutation abolishes the ability of the D257A/N258A NK2 to promote Met-dimerization as compared to Fig. 4_A_.
Similar articles
- Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution.
Ultsch M, Lokker NA, Godowski PJ, de Vos AM. Ultsch M, et al. Structure. 1998 Nov 15;6(11):1383-93. doi: 10.1016/s0969-2126(98)00138-5. Structure. 1998. PMID: 9817840 - Crystal structures of NK1-heparin complexes reveal the basis for NK1 activity and enable engineering of potent agonists of the MET receptor.
Lietha D, Chirgadze DY, Mulloy B, Blundell TL, Gherardi E. Lietha D, et al. EMBO J. 2001 Oct 15;20(20):5543-55. doi: 10.1093/emboj/20.20.5543. EMBO J. 2001. PMID: 11597998 Free PMC article. - A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist.
Tolbert WD, Daugherty J, Gao C, Xie Q, Miranti C, Gherardi E, Vande Woude G, Xu HE. Tolbert WD, et al. Proc Natl Acad Sci U S A. 2007 Sep 11;104(37):14592-7. doi: 10.1073/pnas.0704290104. Epub 2007 Sep 5. Proc Natl Acad Sci U S A. 2007. PMID: 17804794 Free PMC article. - Structural basis of MET receptor dimerization by the bacterial invasion protein InlB and the HGF/SF splice variant NK1.
Niemann HH. Niemann HH. Biochim Biophys Acta. 2013 Oct;1834(10):2195-204. doi: 10.1016/j.bbapap.2012.10.012. Epub 2012 Oct 31. Biochim Biophys Acta. 2013. PMID: 23123275 Review. - Insights into the structure of hepatocyte growth factor/scatter factor (HGF/SF) and implications for receptor activation.
Chirgadze DY, Hepple J, Byrd RA, Sowdhamini R, Blundell TL, Gherardi E. Chirgadze DY, et al. FEBS Lett. 1998 Jun 23;430(1-2):126-9. doi: 10.1016/s0014-5793(98)00558-4. FEBS Lett. 1998. PMID: 9678607 Review.
Cited by
- Monomeric gremlin is a novel vascular endothelial growth factor receptor-2 antagonist.
Grillo E, Ravelli C, Corsini M, Ballmer-Hofer K, Zammataro L, Oreste P, Zoppetti G, Tobia C, Ronca R, Presta M, Mitola S. Grillo E, et al. Oncotarget. 2016 Jun 7;7(23):35353-68. doi: 10.18632/oncotarget.9286. Oncotarget. 2016. PMID: 27174917 Free PMC article. - Semi-synthesis of a HGF/SF kringle one (K1) domain scaffold generates a potent in vivo MET receptor agonist.
Simonneau C, Bérénice Leclercq, Mougel A, Adriaenssens E, Paquet C, Raibaut L, Ollivier N, Drobecq H, Marcoux J, Cianférani S, Tulasne D, de Jonge H, Melnyk O, Vicogne J. Simonneau C, et al. Chem Sci. 2015 Mar 1;6(3):2110-2121. doi: 10.1039/c4sc03856h. Epub 2015 Jan 29. Chem Sci. 2015. PMID: 28717459 Free PMC article. - Transforming growth factor-β1 selectively inhibits hepatocyte growth factor expression via a micro-RNA-199-dependent posttranscriptional mechanism.
Mungunsukh O, Day RM. Mungunsukh O, et al. Mol Biol Cell. 2013 Jul;24(13):2088-97. doi: 10.1091/mbc.E13-01-0017. Epub 2013 May 8. Mol Biol Cell. 2013. PMID: 23657814 Free PMC article. - MET Activation by a Macrocyclic Peptide Agonist that Couples to Biological Responses Differently from HGF in a Context-Dependent Manner.
Miao W, Sakai K, Imamura R, Ito K, Suga H, Sakuma T, Yamamoto T, Matsumoto K. Miao W, et al. Int J Mol Sci. 2018 Oct 12;19(10):3141. doi: 10.3390/ijms19103141. Int J Mol Sci. 2018. PMID: 30322054 Free PMC article. - An engineered dimeric fragment of hepatocyte growth factor is a potent c-MET agonist.
Liu CJ, Jones DS 2nd, Tsai PC, Venkataramana A, Cochran JR. Liu CJ, et al. FEBS Lett. 2014 Dec 20;588(24):4831-7. doi: 10.1016/j.febslet.2014.11.018. Epub 2014 Nov 21. FEBS Lett. 2014. PMID: 25451235 Free PMC article.
References
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. MET, metastasis, motility, and more. Nature Reviews Molecular Cell Biology. 2003;4:915–925. - PubMed
- Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met Signaling pathway in cancer. Clin Cancer Res. 2009;15:2207–2213. - PubMed
- Kirchhofer D, et al. Structural and functional basis of the serine protease-like hepatocyte growth factor β-chain in Met binding and signaling. J Biol Chem. 2004;279:39915–39924. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK066202/DK/NIDDK NIH HHS/United States
- G0800025/MRC_/Medical Research Council/United Kingdom
- G9704528/MRC_/Medical Research Council/United Kingdom
- DK071662/DK/NIDDK NIH HHS/United States
- DK066202/DK/NIDDK NIH HHS/United States
- R01 DK071662/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous