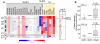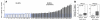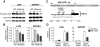Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma - PubMed (original) (raw)
. 2010 Oct 28;116(17):3268-77.
doi: 10.1182/blood-2010-05-282780. Epub 2010 Jul 13.
Stefano Monti, Scott J Rodig, Przemyslaw Juszczynski, Treeve Currie, Evan O'Donnell, Bjoern Chapuy, Kunihiko Takeyama, Donna Neuberg, Todd R Golub, Jeffery L Kutok, Margaret A Shipp
Affiliations
- PMID: 20628145
- PMCID: PMC2995356
- DOI: 10.1182/blood-2010-05-282780
Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma
Michael R Green et al. Blood. 2010.
Abstract
Classical Hodgkin lymphoma (cHL) and mediastinal large B-cell lymphoma (MLBCL) are lymphoid malignancies with certain shared clinical, histologic, and molecular features. Primary cHLs and MLBCLs include variable numbers of malignant cells within an inflammatory infiltrate, suggesting that these tumors escape immune surveillance. Herein, we integrate high-resolution copy number data with transcriptional profiles and identify the immunoregulatory genes, PD-L1 and PD-L2, as key targets at the 9p24.1 amplification peak in HL and MLBCL cell lines. We extend these findings to laser-capture microdissected primary Hodgkin Reed-Sternberg cells and primary MLBCLs and find that programmed cell death-1 (PD-1) ligand/9p24.1 amplification is restricted to nodular sclerosing HL, the cHL subtype most closely related to MLBCL. Using quantitative immunohistochemical methods, we document the association between 9p24.1 copy number and PD-1 ligand expression in primary tumors. In cHL and MLBCL, the extended 9p24.1 amplification region also included the Janus kinase 2 (JAK2) locus. Of note, JAK2 amplification increased protein expression and activity, specifically induced PD-1 ligand transcription and enhanced sensitivity to JAK2 inhibition. Therefore, 9p24.1 amplification is a disease-specific structural alteration that increases both the gene dosage of PD-1 ligands and their induction by JAK2, defining the PD-1 pathway and JAK2 as complementary rational therapeutic targets.
Figures
Figure 1
Chromosome 9p24.1 amplification and increased expression of PD-1 ligands in HL and MLBCL cell lines. (A left) Smoothed chromosome 9p gene copy number estimates for each DLBCL, MLBCL, and HL cell line. The color scale ranges from blue (deletion) to red (amplification). (Right) GISTIC Q values (for all cell lines) for the 9p24.1 amplification that includes the PD-L1/PD-L2 loci. (B) PD-L1 and PD-L2 transcript abundance in the DLBCL, MLBCL, and HL cell lines was assessed by transcriptional profiling and represented in box plots.
Figure 2
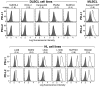
9p24.1 amplification and PD-1 ligand cell-surface expression in HL and MLBCL cell lines. Flow cytometric analysis of cell-surface PD-1 ligand (PD-L1 and PD-L2) expression in DLBCL, MLBCL, and HL cell lines (PD-L1 or PD-L2, open black lines; isotype controls, solid gray histograms). DLBCL and HL cell lines are arranged according to PD-1 ligand copy number (normal to high, left to right).
Figure 3
PD-1 ligand amplification and overexpression in primary HL. (A) Laser-capture microdissection (LCM) of primary HL Reed-Sternberg RS cells. (Ai) RS cells were identified by CD30 staining, (Aii) selected by laser (Arcturus PixCell II), (Aiii) removed from surrounding nonneoplastic tissue, resulting in (Aiv) highly enriched RS cell specimens. (B) qPCR-based DNA copy number analysis of PD-L1 in LCM primary RS cells isolated from 7 MCHLs and 16 NSHLs. Data are means (± SD) of triplicate qPCR reactions performed on pooled DNA samples from RS cells or normal tissue from each slide, repeated for 3 slides per sample. (C left) Quantitative IHC of PD-L1 in 3 representative cases (8, 15, and 22) from panel B. (C right) Quantitative analysis of PD-L1 immunostaining in 150 individual RS cells from each of these cases (8, 15, and 22).
Figure 4
PD-1 ligand amplification and overexpression in primary MLBCL. (A) qPCR-based DNA copy number analysis of PD-L1 in 41 primary MLBCLs. (B) RT-qPCR analysis of PD-L1 and PD-L2 transcript abundance in the same series of primary MLBCLs. Transcript abundance in tumors with normal or increased 9p24.1 is represented in box blots.
Figure 5
JAK2 amplification, expression, and activity. (A) GISTIC analysis (for all cell lines) of 9p24.1, PD-L1 and PD-L2, in the amplification peak and JAK2 in the broader amplification region. (B) JAK2 transcript abundance in HL and DLBCL cell lines and the single MLBCL cell line (left) and primary MLBCLs with 9p24.1 (PD-L1) amplification or normal copy number (right). (C) Western blot analysis of phospho- and total JAK2 in HL cell lines arranged according to 9p24.1 copy number (normal to high, left to right, as in Figure 2) and the single MLBCL cell line. (D) Intracellular phosphoflow cytometric analysis of phosphorylated STAT1 in HL and MLBCL cell lines in panel C.
Figure 6
Chemical inhibition of JAK2 decreases PD-1 ligand transcription. (A) Western analysis of phosphoJAK2 in HL cell lines (L428 and SUPHD1) treated with the increasing doses (2.5-10μm) of the specific JAK2 inhibitor, SD-1029. (B) RT-qPCR analysis of PD-L1 transcript abundance in the cell lines treated with SD-1029. Data are means (± SD) of triplicate measurements from the representative experiment shown in panel A. (C) The PD-L1 promoter regulatory module. STAT-responsive (ISRE) element and additional degenerate STAT-binding sites are indicated. This region was cloned into a pGL-luciferase vector (pGL3-PD-L1p). (D) Analysis of pGL3-PD-L1p luciferase activity in L428 and SUPHD1 HL cells treated with SD-1029 or vehicle. Data are means (± SD) of triplicate measurements from a representative experiment. Experiments in panels B and D were performed 3 times.
Figure 7
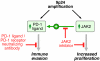
9p24.1 amplification targets, consequences, and associated treatment options. 9p24.1 amplification increases PD-1 ligand (PD-L1 and PD-L2) and JAK2 copy numbers, augments JAK2/STAT1 activity, induces PD-1 ligand expression, and stimulates HL proliferation.
Similar articles
- Selective JAK2 inhibition specifically decreases Hodgkin lymphoma and mediastinal large B-cell lymphoma growth in vitro and in vivo.
Hao Y, Chapuy B, Monti S, Sun HH, Rodig SJ, Shipp MA. Hao Y, et al. Clin Cancer Res. 2014 May 15;20(10):2674-83. doi: 10.1158/1078-0432.CCR-13-3007. Epub 2014 Mar 7. Clin Cancer Res. 2014. PMID: 24610827 Free PMC article. - PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome.
Roemer MG, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, Connelly CF, Sun HH, Daadi SE, Freeman GJ, Armand P, Chapuy B, de Jong D, Hoppe RT, Neuberg DS, Rodig SJ, Shipp MA. Roemer MG, et al. J Clin Oncol. 2016 Aug 10;34(23):2690-7. doi: 10.1200/JCO.2016.66.4482. Epub 2016 Apr 11. J Clin Oncol. 2016. PMID: 27069084 Free PMC article. - The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma.
Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, Aguiar RC, Li S, Salles G, Berger F, Jing W, Pinkus GS, Habermann T, Dalla-Favera R, Harris NL, Aster JC, Golub TR, Shipp MA. Savage KJ, et al. Blood. 2003 Dec 1;102(12):3871-9. doi: 10.1182/blood-2003-06-1841. Epub 2003 Aug 21. Blood. 2003. PMID: 12933571 - Signaling pathways and immune evasion mechanisms in classical Hodgkin lymphoma.
Liu WR, Shipp MA. Liu WR, et al. Hematology Am Soc Hematol Educ Program. 2017 Dec 8;2017(1):310-316. doi: 10.1182/asheducation-2017.1.310. Hematology Am Soc Hematol Educ Program. 2017. PMID: 29222272 Free PMC article. Review. - Signaling pathways and immune evasion mechanisms in classical Hodgkin lymphoma.
Liu WR, Shipp MA. Liu WR, et al. Blood. 2017 Nov 23;130(21):2265-2270. doi: 10.1182/blood-2017-06-781989. Blood. 2017. PMID: 29167175 Free PMC article. Review.
Cited by
- Long-term Benefit of PD-L1 Blockade in Lung Cancer Associated with JAK3 Activation.
Van Allen EM, Golay HG, Liu Y, Koyama S, Wong K, Taylor-Weiner A, Giannakis M, Harden M, Rojas-Rudilla V, Chevalier A, Thai T, Lydon C, Mach S, Avila AG, Wong JA, Rabin AR, Helmkamp J, Sholl L, Carter SL, Oxnard G, Janne P, Getz G, Lindeman N, Hammerman PS, Garraway LA, Hodi FS, Rodig SJ, Dranoff G, Wong KK, Barbie DA. Van Allen EM, et al. Cancer Immunol Res. 2015 Aug;3(8):855-63. doi: 10.1158/2326-6066.CIR-15-0024. Epub 2015 May 26. Cancer Immunol Res. 2015. PMID: 26014096 Free PMC article. - Human Herpesvirus and the Immune Checkpoint PD-1/PD-L1 Pathway: Disorders and Strategies for Survival.
Murata T. Murata T. Microorganisms. 2021 Apr 8;9(4):778. doi: 10.3390/microorganisms9040778. Microorganisms. 2021. PMID: 33917804 Free PMC article. Review. - PD-L1 Detection in Tumors Using [(64)Cu]Atezolizumab with PET.
Lesniak WG, Chatterjee S, Gabrielson M, Lisok A, Wharram B, Pomper MG, Nimmagadda S. Lesniak WG, et al. Bioconjug Chem. 2016 Sep 21;27(9):2103-10. doi: 10.1021/acs.bioconjchem.6b00348. Epub 2016 Aug 9. Bioconjug Chem. 2016. PMID: 27458027 Free PMC article. - Molecular Pathogenesis of Hodgkin Lymphoma: Past, Present, Future.
Bienz M, Ramdani S, Knecht H. Bienz M, et al. Int J Mol Sci. 2020 Sep 10;21(18):6623. doi: 10.3390/ijms21186623. Int J Mol Sci. 2020. PMID: 32927751 Free PMC article. Review. - Ipilimumab, nivolumab, and brentuximab vedotin combination therapies in patients with relapsed or refractory Hodgkin lymphoma: phase 1 results of an open-label, multicentre, phase 1/2 trial.
Diefenbach CS, Hong F, Ambinder RF, Cohen JB, Robertson MJ, David KA, Advani RH, Fenske TS, Barta SK, Palmisiano ND, Svoboda J, Morgan DS, Karmali R, Sharon E, Streicher H, Kahl BS, Ansell SM. Diefenbach CS, et al. Lancet Haematol. 2020 Sep;7(9):e660-e670. doi: 10.1016/S2352-3026(20)30221-0. Lancet Haematol. 2020. PMID: 32853585 Free PMC article. Clinical Trial.
References
- Kuppers R. The biology of Hodgkin's lymphoma. Nat Rev Cancer. 2009;9:15–27. - PubMed
- Savage K, Monti S, Kutok J, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102(12):3871–3879. - PubMed
- Gaulard P, Harris NL, Pileri SA, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Geneva, Switzerland: WHO Press; 2008. Primary mediastinal (thymic) large B-cell lymphoma. pp. 175–176.
- Stein H, von Wasielewski R, Poppema S, MacLennan KA, Guenovva M. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Geneva, Switzerland: WHO Press; 2008. Nodular sclerosis classical Hodgkin lymphoma. pp. 249–250.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous