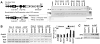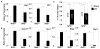Regulation of notch1 signaling by nrf2: implications for tissue regeneration - PubMed (original) (raw)
Regulation of notch1 signaling by nrf2: implications for tissue regeneration
Nobunao Wakabayashi et al. Sci Signal. 2010.
Abstract
The Keap1-Nrf2-ARE signaling pathway elicits an adaptive response for cell survival after endogenous and exogenous stresses, such as inflammation and carcinogens, respectively. Keap1 inhibits the transcriptional activation activity of Nrf2 (p45 nuclear factor erythroid-derived 2-related factor 2) in unstressed cells by facilitating its degradation. Through transcriptional analyses in Keap1- or Nrf2-disrupted mice, we identified interactions between the Keap1-Nrf2-ARE and the Notch1 signaling pathways. We found that Nrf2 recognized a functional antioxidant response element (ARE) in the promoter of Notch1. Notch1 regulates processes such as proliferation and cell fate decisions. We report a functional role for this cross talk between the two pathways and show that disruption of Nrf2 impeded liver regeneration after partial hepatectomy and was rescued by reestablishment of Notch1 signaling.
Conflict of interest statement
Competing interests: There may be MTAs related to the new mouse constructs, per the policy of Johns Hopkins University.
Figures
Fig. 1. Differential expression of Notch1 and related genes in wild-type (Nrf2+/+) and Nrf2-null (_Nrf2_−/−) MEFs
A. Semi-quantitative RT- PCR for Notch1, Notch1 effector genes (Hes-1, Herp1, Herp2, Nrarp, p21) and Notch1 ligand-encoding gene, Jag1. The abundance of the transcript for Tata binding protein (TBP) served as a loading control. Data are quantified as the ratio of the expression of the indicated genes in Nrf2 null/wild type. Values are the means of 3 independent experiments ± S.E. Expression of all effector genes, except Jag1, was significantly lower in the Nrf2 null MEF (p < 0.01; t- test). B. Real-time RT-PCR confirmation of changes in Notch1 expression in wild-type versus _Nrf2_-null MEF. The loss of induction of Gsta1, a Nrf2-responsive gene, confirmed the disruption in Nrf2 activity. MEF cultures were treated with 2.5 μM sulforaphane or vehicle 24 h before harvesting cells for RNA isolation. Values are mean ± S.E. * p < 0.05; t-test (n=3). C. RT- PCR analyses of Notch1 and other Nrf2 target gene expression in the livers from 8-week old _Nrf2_-null, wild-type, and various Keap1 and Nrf2 compound-disrupted mice. Hypoxanthine guanine phosphoribosyl transferase (Hprt) served as the loading control. For quantitiation see supplemental Fig. 1B and 1C.
Fig. 2. Analysis of the proximal promoter of murine Notch1 in MEFs
A ~2-kb portion of the promoter of Notch1 was isolated from murine liver and ligated into a luciferase reporter vector (−1640 Notch1-Luc) to monitor its activity. A. Distribution of putative ARE motifs. B. Effect of the presence of Nrf2 and sulforaphane (2.5 μM) treatment on −1640 _Notch1_-Luc reporter gene activity in MEFs. Values are mean ± S.E. (n=3). * P < 0.05. Data are shown as the relative luciferase activity (RLA) compared to that in vector (pCMV Mock )-transfected cells in the absence of sulforaphane, which was set at a value of 1.0. C. Serial deletion assays of the −1640 _Notch1_-Luc reporter with relative luciferase activities in MEF (left) and P19 EC (right) cells shown. In both analyses, the activities from the −78 construct were set at a value of 1.0. Black ellipses show the presence of putative AREs. Values are mean ± S.D. (n=3). Luciferase activities were normalized by measuring the Renilla luciferase activity from a cotransfected reporter vector. * P < 0.05. D. Effect of site-directed mutagenesis on the enhancer capacity of the 1-ARE motif in MEFs. Cells were either transfected with the −206 _Notch1_-Luc truncated promoter construct (p-206 Luc), which contained the 1-ARE, or with a form with point mutations in the 1-ARE (p-206 mARE Luc). RLA was based on the activity from each −206 Luc reporter construct when transfected with the vector pCMV Mock, which was set at a value of 1.0. Values are mean ± S.D. * p < 0.05. E. Recombinant Nrf2 and MafK form a complex with _Notch1_-1-ARE, which can be competed away by the _Gsta1_-ARE but not mutant 1-ARE. Left panels show negative controls for the analysis. Right panels show EMSA in the presence of the ARE. F. Binding of nuclear factors from MEFs to the 1-ARE of Notch1 can be competed away with the ARE of the mouse Gsta1 promoter region (table S1), but not by mutant 1-ARE of Notch1 (mARE). A 25-fold molar excess of each competitor was used. G. The binding complex can be supershifted with an antibody to Nrf2, but not with an antibody to Nrf1 or with normal rabbit IgG. The arrowhead indicates the specific protein complex with the 1-ARE probe and the arrow indicates the supershifted band. H. ChIP assay using wild-type and _Nrf2_−/− cells. No DNA and IgG reactions were the controls for PCR and immunoprecipitation steps, respectively. The 1-ARE in the Notch1 gene regulatory region was detected only in the presence of wild-type cell nuclear extract and the antibody to Nrf2. The Gsta1 promoter region, which has a functional ARE, served as a positive control.
Fig. 3. The timing-specific induction of Notch1 by Nrf2 is shown with RosaCreER/CreER:: Keap1flox/flox (RCKF) MEF cells
A. Scheme of the genetic alleles present within the mouse constructs utilized: The mutant cell line carrying the floxed Keap1 gene (top) and the expected disrupted allele (bottom). RosaCreER/CreER MEF cells (RC1 and RC2), as well as RCKF1 and RCKF2 cells, were treated with 1 μM 4-OHT for 3 days with repetitive treatments each 24 hours. D, disrupted gene; Un, undisrupted gene. B. Treatment with 4-OHT induces Notch1 and Nqo1 gene expression in RCKF MEF cells, as shown by RT-PCR. The magnitude of the expression of each gene was normalized by the expression of the housekeeping gene Hprt using the total RNA derived from both RC1 and RCKF1 MEF (right panel). Gene expression from vehicle treated samples in each cell was set at a value of 1.0. Values are mean ± S.D. * p < 0.01 (n=3, one-way analysis of variance. C. Immunoblotting of Nrf2 in the RCKF MEF cell extracts shows that Nrf2 accumulates in cells following treatment with 4-OHT.
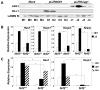
Fig. 4. In MEFs, Notch1 signaling is dependent upon Nrf2 genotype
A. The abundance of Notch1 signaling ligands (DLL1 or JAG1) in independent lines of stably transfected HEK293 cells as shown by immunoblot analysis with JAG1 or DLL1 specific antibodies (sc-8303, sc-9102). Probing the blots with a Lamin B1 antibody (sc-6216) served as the loading control. M1-4, mock; D1-4, DLL1; J1-4, JAG1. B. Effect of Nrf2 genotype on Notch1 signaling in MEFs co-cultured with JAG1-expressing HEK293 (J2) cells. C. Time course showing Notch1 signaling in MEF cells co-cultured with DLL1 expressing HEK293 (D2) cells. Gene expression in B and C was examined by real time reverse transcriptase PCR. 18S ribosomal RNA expression was used for normalization. The expression for each gene in Nrf2+/+ MEF was set at a value of 1.0. Values are mean ± S.D. (n=3). *P< 0.05, one-way analysis of variance.
Fig. 5. Reduced basal Notch1 signaling in _Nrf2_-null mice delays liver regeneration
A. Jag1 and Dll1 expression in whole liver was analyzed by real-time reverse transcriptase PCR as ligans of Notch1 signaling 3 hr after 2/3 partial hepatectomy. Hprt expression was used for normalization. B. Hes1 expression was monitored by real-time reverse transcriptase PCR as an index of Notch1 signaling 6 hr after 2/3 partial hepatectomy. Albumin expression was used for normalization. The ratio of hepatic gene of interest to albumin gene expression in sham-operated Nrf2+/+ mice was set at a value of 1.0. Values are mean ± S.D., n= 3, * p < 0.05, one-way analysis of variance. C. Relative liver weights from _Nrf2_−/− and wild-type mice (age and total body weight-matched) 3 days after 2/3 partial hepatectomy (PH). Values are mean ± S.D. * p < 0.01, one-way analysis of variance. White boxes in A, B and C represent sham-operated animals; black boxes are partial hepatectomy animals.
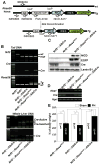
Fig. 6. NICD restores liver regeneration in _Nrf2_-null mice
A. Scheme for hepatocyte-specific NICD expression. Arrows show primers used for genotyping. Sequences are described in table S2. B. Demonstration of homologous recombination only in genomic DNA isolated from the livers of Nrf2_−/−::RosaNICD/−::AlbCre_ mice. Wt, Tg, mut and Cre stand for wild-type locus, transgenic rosa 26 locus, Nrf2 disrupted allele and AlbCre recombinase insertion alle. C. Immunoblots of NICD and EGFP demonstrating that the proteins were present only in the liver of Nrf2_−/−::RosaNICD/−::AlbCre_ mice. Lamin B1 served as the control for protein loading and was detected with an antibody. D. Hepatic expression of Hes1 in Nrf2_−/−::RosaNICD/−, Nrf2_−/−::AlbCre, and Nrf2_−/−::RosaNICD/−::AlbCre_ mice. The ratios of hepatic Hes1 expression are 0.58±0.13(SD), 4.31±0.72(SD) in Nrf2_−/−::RosaNICD/− and Nrf2_−/−::RosaNICD/−::AlbCre, respectively_. Hes1_ expression in Nrf2_−/−::AlbCre_ was set at a value of 1.0. n=3, p<0.01, one-way analysis of variance. E. Regenerative recovery of liver mass in Nrf2_−/−::RosaNICD/−, Nrf2_−/−::AlbCre and Nrf2_−/−::RosaNICD/−::AlbCre_ mice. Values are mean ± S.D., * p < 0.01, one-way analysis of variance. White box, sham-operated animals; black boxes, partial hepatectomy animals.
Similar articles
- When NRF2 talks, who's listening?
Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. Wakabayashi N, et al. Antioxid Redox Signal. 2010 Dec 1;13(11):1649-63. doi: 10.1089/ars.2010.3216. Epub 2010 Jul 9. Antioxid Redox Signal. 2010. PMID: 20367496 Free PMC article. Review. - Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice.
Yates MS, Tran QT, Dolan PM, Osburn WO, Shin S, McCulloch CC, Silkworth JB, Taguchi K, Yamamoto M, Williams CR, Liby KT, Sporn MB, Sutter TR, Kensler TW. Yates MS, et al. Carcinogenesis. 2009 Jun;30(6):1024-31. doi: 10.1093/carcin/bgp100. Epub 2009 Apr 21. Carcinogenesis. 2009. PMID: 19386581 Free PMC article. - Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis.
Kong X, Thimmulappa R, Craciun F, Harvey C, Singh A, Kombairaju P, Reddy SP, Remick D, Biswal S. Kong X, et al. Am J Respir Crit Care Med. 2011 Oct 15;184(8):928-38. doi: 10.1164/rccm.201102-0271OC. Epub 2011 Jul 28. Am J Respir Crit Care Med. 2011. PMID: 21799073 Free PMC article. - Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2 x Keap1 x Cul3 complex and recruiting Nrf2 x Maf to the antioxidant response element enhancer.
He X, Chen MG, Lin GX, Ma Q. He X, et al. J Biol Chem. 2006 Aug 18;281(33):23620-31. doi: 10.1074/jbc.M604120200. Epub 2006 Jun 19. J Biol Chem. 2006. PMID: 16785233 - Nrf2 protects against airway disorders.
Cho HY, Kleeberger SR. Cho HY, et al. Toxicol Appl Pharmacol. 2010 Apr 1;244(1):43-56. doi: 10.1016/j.taap.2009.07.024. Epub 2009 Jul 29. Toxicol Appl Pharmacol. 2010. PMID: 19646463 Review.
Cited by
- Nrf2 participates in regulating maternal hepatic adaptations to pregnancy.
Zou Y, Hu M, Bao Q, Chan JY, Dai G. Zou Y, et al. J Cell Sci. 2013 Apr 1;126(Pt 7):1618-25. doi: 10.1242/jcs.118109. Epub 2013 Feb 15. J Cell Sci. 2013. PMID: 23418358 Free PMC article. - Sex differences in resilience to ferroptosis underlie sexual dimorphism in kidney injury and repair.
Ide S, Ide K, Abe K, Kobayashi Y, Kitai H, McKey J, Strausser SA, O'Brien LL, Tata A, Tata PR, Souma T. Ide S, et al. Cell Rep. 2022 Nov 8;41(6):111610. doi: 10.1016/j.celrep.2022.111610. Cell Rep. 2022. PMID: 36351395 Free PMC article. - Nrf2 protects stellate cells from Smad-dependent cell activation.
Prestigiacomo V, Suter-Dick L. Prestigiacomo V, et al. PLoS One. 2018 Jul 20;13(7):e0201044. doi: 10.1371/journal.pone.0201044. eCollection 2018. PLoS One. 2018. PMID: 30028880 Free PMC article. - Redox Signaling by Reactive Electrophiles and Oxidants.
Parvez S, Long MJC, Poganik JR, Aye Y. Parvez S, et al. Chem Rev. 2018 Sep 26;118(18):8798-8888. doi: 10.1021/acs.chemrev.7b00698. Epub 2018 Aug 27. Chem Rev. 2018. PMID: 30148624 Free PMC article. Review. - Nrf2 Weaves an Elaborate Network of Neuroprotection Against Stroke.
Jiang S, Deng C, Lv J, Fan C, Hu W, Di S, Yan X, Ma Z, Liang Z, Yang Y. Jiang S, et al. Mol Neurobiol. 2017 Mar;54(2):1440-1455. doi: 10.1007/s12035-016-9707-7. Epub 2016 Feb 5. Mol Neurobiol. 2017. PMID: 26846360 Review.
References
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. - PubMed
- Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266:11632–11639. - PubMed
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. - PubMed
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA94076/CA/NCI NIH HHS/United States
- T32 CA009110/CA/NCI NIH HHS/United States
- R01 CA140492/CA/NCI NIH HHS/United States
- P30 ES03819/ES/NIEHS NIH HHS/United States
- P30 ES003819/ES/NIEHS NIH HHS/United States
- R01 CA094076/CA/NCI NIH HHS/United States
- R01 HL081205/HL/NHLBI NIH HHS/United States
- T32 ES007141/ES/NIEHS NIH HHS/United States
- T32 ES07141/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases