The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies - PubMed (original) (raw)
The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies
Patrick D Schloss. PLoS Comput Biol. 2010.
Abstract
Pyrosequencing of PCR-amplified fragments that target variable regions within the 16S rRNA gene has quickly become a powerful method for analyzing the membership and structure of microbial communities. This approach has revealed and introduced questions that were not fully appreciated by those carrying out traditional Sanger sequencing-based methods. These include the effects of alignment quality, the best method of calculating pairwise genetic distances for 16S rRNA genes, whether it is appropriate to filter variable regions, and how the choice of variable region relates to the genetic diversity observed in full-length sequences. I used a diverse collection of 13,501 high-quality full-length sequences to assess each of these questions. First, alignment quality had a significant impact on distance values and downstream analyses. Specifically, the greengenes alignment, which does a poor job of aligning variable regions, predicted higher genetic diversity, richness, and phylogenetic diversity than the SILVA and RDP-based alignments. Second, the effect of different gap treatments in determining pairwise genetic distances was strongly affected by the variation in sequence length for a region; however, the effect of different calculation methods was subtle when determining the sample's richness or phylogenetic diversity for a region. Third, applying a sequence mask to remove variable positions had a profound impact on genetic distances by muting the observed richness and phylogenetic diversity. Finally, the genetic distances calculated for each of the variable regions did a poor job of correlating with the full-length gene. Thus, while it is tempting to apply traditional cutoff levels derived for full-length sequences to these shorter sequences, it is not advisable. Analysis of beta-diversity metrics showed that each of these factors can have a significant impact on the comparison of community membership and structure. Taken together, these results urge caution in the design and interpretation of analyses using pyrosequencing data.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures
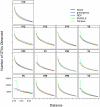
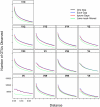
Figure 1. The number of OTUs observed as a function of genetic distance for various regions within the 16S rRNA gene when using different sequence alignments.
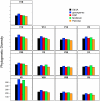
Figure 2. The phylogenetic diversity observed for different regions within the 16S rRNA gene when using different alignments.
Phylogenetic diversity was measured by calculating the total branch length for a phylogenetic tree.
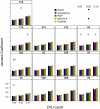
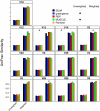
Figure 3. The Jaccard coefficient calculated between two mock communities (described in Materials & Methods) for different OTU definitions and alignments.
Each bar represents the average coefficient value for 100 randomized partitionings of the data. Within the same OTU cutoff, alignment strategies with the same symbol and regions with the same letter were not significantly different from each other.
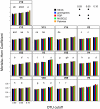
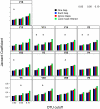
Figure 4. The Morisita-Horn coefficient calculated between two mock communities (described in Materials & Methods) using different OTU definitions and alignments.
Each bar represents the average coefficient value for 100 randomized partitionings of the data. Within the same OTU cutoff, alignment strategies with the same symbol and regions with the same letter were not significantly different from each other.
Figure 5. Unweighted and weighted UniFrac similarity values calculated between two mock communities (described in Materials & Methods) using different alignments.
Each bar represents the average coefficient value for 100 randomized partitionings of the data. Within the same UniFrac approach, alignment strategies with the same symbol and regions with the same letter were not significantly different from each other.
Figure 6. The number of OTUs observed as a function of genetic distance for various regions within the 16S rRNA gene when using different methods of calculating distances and masking sequences.
Figure 7. The phylogenetic diversity observed for different regions within the 16S rRNA gene when using different methods of calculating distances and masking sequences.
Phylogenetic diversity was measured by calculating the total branch length for a phylogenetic tree.
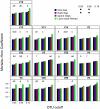
Figure 8. Jaccard similarity values calculated between two mock communities (described in Materials & Methods) for different OTU definitions, methods of calculating distances, and masking sequences.
Each bar represents the average coefficient value for 100 randomized partitionings of the data. Within the same OTU cutoff, regions with the same letter were not significantly different from each other; for each OTU cutoff all distance calculation methods were significantly different from each other.
Figure 9. The Morisita-Horn coefficient calculated between two mock communities (described in Materials & Methods) using different OTU definitions, methods of calculating distances, and masking sequences.
Each bar represents the average coefficient value for 100 randomized partitionings of the data. Within the same OTU cutoff, distance calculation methods with the same symbol and regions with the same letter were not significantly different from each other.
Figure 10. Unweighted and weighted UniFrac similarity values calculated between two mock communities (described in Materials & Methods) using different methods of calculating distances and masking sequences.
Each bar represents the average coefficient value for 100 randomized partitionings of the data. Within the same UniFrac method, regions with the same letter were not significantly different from each other; for both UniFrac methods the distance calculation methods were all significantly different from each other.
References
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous