Promoter hypomethylation up-regulates CD147 expression through increasing Sp1 binding and associates with poor prognosis in human hepatocellular carcinoma - PubMed (original) (raw)
Promoter hypomethylation up-regulates CD147 expression through increasing Sp1 binding and associates with poor prognosis in human hepatocellular carcinoma
Ling-Min Kong et al. J Cell Mol Med. 2011 Jun.
Abstract
CD147 is a transmembrane glycoprotein overexpressed in human hepatocellular carcinoma (HCC) which could promote HCC progression and metastasis. Promoter methylation is one of the most important processes in gene regulation. In this study, we aim to investigate CD147 promoter methylation status and the correlation with clinicopathological features and prognosis in HCC. CD147 promoter methylation statuses and expression levels in normal and HCC cell lines and 54 paired HCC and adjacent non-tumour (ANT) tissues were, respectively, examined by bisulphite genomic sequencing, methylation-specific PCR, real-time RT-PCR, Western blot and immunohistochemistry. The correlations of promoter methylation statuses with CD147 expression level and the clinicopathological features were statistically analysed in HCC patients. Significantly higher expression of CD147 and significantly lower promoter methylation level were observed in HCC cell lines compared to normal cell lines and tissues control. In vivo and in vitro analysis indicated that demethylation with 5-Aza-2'-deoxycytidine led to increased CD147 expression through enhancing Sp1 binding affinity, and methylation with methyltransferase reduced CD147 transcriptional activity through interfering Sp1 binding. CD147 promoter methylation level in HCC tissues (22.22%) was lower than that in ANT tissues (46.30%; P < 0.05). Within HCC tissues, a significant inverse correlation was observed between CD147 expression and methylation level (r=-0.615). Moreover, HCC patients with unmethylated CD147 promoter had a significantly higher recurrence rate (88.1%versus 58.3%; P < 0.05) and death rate (83.3%versus 50.0%; P < 0.05) than patients with methylated CD147 promoter. In conclusions, promoter hypomethylation up-regulates CD147 expression primarily through increasing Sp1 binding and associates with poor prognosis in HCC patients.
© 2011 The Authors Journal of Cellular and Molecular Medicine © 2011 Foundation for Cellular and Molecular Medicine/Blackwell Publishing Ltd.
Figures
Fig 1
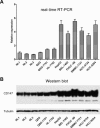
Up-expression of CD147 in HCC cell lines. (A) mRNA expressions of CD147 gene were detected by real-time quantitative RT-PCR in HCC cell lines, normal cell lines and normal liver tissues. GAPDH gene was used as an internal control. (B) CD147 protein expression was determined by Western blot with tubulin as an internal control.
Fig 2
DNA Methylation profile of the CpG island at the CD147 promoter region. (A) Map of predicted CpG islands. One CpG island located at −340 to +37 was identified in CD147 promoter (+1, transcriptional start site; ATG, translational starting site). GC percentage and observed/expected CpG were calculated using the CPGPLOT program. (B) Schematic representation of CpG island. The 46 CG dinucleotides were denoted as grey shadow. CpG sites were numbered from 1 to 46 relative to the transcription start site (+1 indicated with an arrow). Underlined region indicated Sp1 binding sites. (C) Sequences of the CD147 promoter region after bisulphite modification were analysed in normal and cancer cell lines. Black circle, methylated cytosine; white circle, unmethylated cytosine in the dinucleotide CpG.
Fig 3
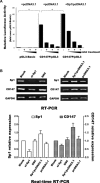
The involvement of Sp1 in regulating CD147 expression. (A) Analysis for transcription activity of CD147 promoter and the regulation role of Sp1 in HEK-293 cell by the co-transfection of luciferase reporter plasmid (CD147P/pGL3 or pGL3-Basic) and expression vector (pcDNA3.1 or Sp1/pcDNA3.1). Transfected cells were either untreated or treated with increasing concentration (0.5, 1, 5 μM) of mithramycin A. The luciferase activity value of each sample was first normalized for transfection efficiency by co-transfection with the pRL-TK plasmid. The transcriptional activity of promoter construct was shown as the luciferase activity relative to that of the pGL3-Basic vector (a promoter-less vector) and shown as the mean ± S.D. for three independent experiments. *P < 0.05. (B) The involvement of Sp1 in regulating CD147 expression. HepG2 cell was transfected with siRNA targeting Sp1 and Sp1 expression vector, respectively. Upper panel, regular RT-PCR. Lower panel, real-time quantitative RT-PCR. Control siRNA was used as negative control.
Fig 4
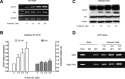
Promoter hypomethylation up-regulated CD147 expression through increasing Sp1 binding in vivo. (A, B and C) The mRNA and protein expression of CD147 and Sp1 detected by regular RT-PCR, real-time quantitative RT-PCR and Western blot in QZG cell treated with different concentration of 5-Aza-dC. (D) Demethylation increased the binding of Sp1 to the CD147 promoter in vivo. ChIP assay using antibody against Sp1 was performed in before and after 5-Aza-dC treatment QZG cell. The normal rabbit IgG was used as a negative control and Input indicates 5% input DNA, a positive amplification control.
Fig 5
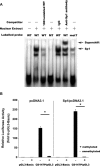
Methylation status interfered the Sp1 binding to the CD147 gene promoter and inhibited CD147 transcriptional activity in vitro. (A) Methylation status interfered the Sp1 binding to the CD147 gene promoter in vitro. The binding of Sp1 to the CD147 gene promoter was determined by electrophoretic mobility shift analysis (EMSA). By using biotin-labelled 30-bp double-stranded oligonucleotides containing wild, mutated or methylated Sp1-binding sites as probes, EMSAs were performed with the same amount of nuclear extracts from HepG2 cells, and the products were separated on a 5% polyacrylamide gel (lanes 2–7). Lane 1, free probe; lane 2, biotin-labelled wild-type Sp1 consensus oligonucleotides were mixed with nuclear proteins; lane 3, the same reaction was performed as that in lane 2, except for the presence of a 100-fold excess of unlabelled wild-type Sp1 consensus oligonucleotides as a competitor; lanes 4, binding assays of biotin-labelled mutant-type Sp1 consensus oligonucleotides mixed with nuclear proteins; lanes 5–6, 1 μg each of IgG and anti-Sp1 antibody were added to the binding reaction mixtures with biotin-labelled wild-type probe; lane 7, binding assays of biotin-labelled methylated Sp1 consensus oligonucleotides mixed with nuclear proteins. (B) Analysis for the effect of in vitro DNA methylation on the CD147 promoter activity through the transfection of HEK-293 cells with methylated CpG reporter constructs. CD147P/pGL3 treated or untreated with SssI methylase was co-transfected with the pcDNA3.1 or Sp1/pcDNA3.1 into HEK-293 cells with pGL3-Basic as control. The relative luciferase activity was denoted as above-mentioned method and also expressed as the mean ± S.D. for three independent experiments. *P < 0.05.
Fig 6
Analysis of methylation status in CD147 promoter and CD147 expression level in HCC and ANT tissues. (A) Representative methylation profiles of CpG island in CD147 promoter in HCC and ANT tissues detected by BGS. Open and closed circles indicate unmethylated and methylated CpG sites, respectively. The percentage of methylated CpG sites in all sequenced CpG sites is shown in parentheses. (B) Representative results of MS-PCR analysis in HCC tumour tissues (T) and ANT tissues (N). M: methylation; U: unmethylation. (C) Immunohistochemical staining analysis for CD147 expression in HCC tissues. Top, positive CD147 immunostaining. Bottom, negative CD147 immunostaining. Scale bars, 50 μm. (D) Analysis for correlation of CD147 expression and its promoter metheylation in HCC tissues by calculating Spearman’s ρ method.
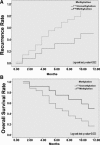
Fig 7
Kaplan–Meier recurrence and overall survival estimates based on the methylation status of CD147 promoter for all HCC patients. (A) Kaplan–Meier recurrence curve. (B) Kaplan–Meier overall survival curve.
Similar articles
- Methylation of Tip30 promoter is associated with poor prognosis in human hepatocellular carcinoma.
Lu B, Ma Y, Wu G, Tong X, Guo H, Liang A, Cong W, Liu C, Wang H, Wu M, Zhao J, Guo Y. Lu B, et al. Clin Cancer Res. 2008 Nov 15;14(22):7405-12. doi: 10.1158/1078-0432.CCR-08-0409. Clin Cancer Res. 2008. PMID: 19010857 - Methylation of IRAK3 is a novel prognostic marker in hepatocellular carcinoma.
Kuo CC, Shih YL, Su HY, Yan MD, Hsieh CB, Liu CY, Huang WT, Yu MH, Lin YW. Kuo CC, et al. World J Gastroenterol. 2015 Apr 7;21(13):3960-9. doi: 10.3748/wjg.v21.i13.3960. World J Gastroenterol. 2015. PMID: 25852282 Free PMC article. - ZBP-89 and Sp1 contribute to Bak expression in hepatocellular carcinoma cells.
Kong X, Xu P, Cai WJ, Wang HG, Li BB, Huang GL, He ZW, Chen G, Ye CG. Kong X, et al. BMC Cancer. 2018 Apr 13;18(1):419. doi: 10.1186/s12885-018-4349-y. BMC Cancer. 2018. PMID: 29653560 Free PMC article. - Clinical implications of DNA methylation in hepatocellular carcinoma.
Sceusi EL, Loose DS, Wray CJ. Sceusi EL, et al. HPB (Oxford). 2011 Jun;13(6):369-76. doi: 10.1111/j.1477-2574.2011.00303.x. Epub 2011 Mar 29. HPB (Oxford). 2011. PMID: 21609368 Free PMC article. Review. - Role of CD147 in the development and diagnosis of hepatocellular carcinoma.
Huang D, Rao D, Jin Q, Lai M, Zhang J, Lai Z, Shen H, Zhong T. Huang D, et al. Front Immunol. 2023 Apr 6;14:1149931. doi: 10.3389/fimmu.2023.1149931. eCollection 2023. Front Immunol. 2023. PMID: 37090718 Free PMC article. Review.
Cited by
- The biological function and clinical utilization of CD147 in human diseases: a review of the current scientific literature.
Xiong L, Edwards CK 3rd, Zhou L. Xiong L, et al. Int J Mol Sci. 2014 Sep 29;15(10):17411-41. doi: 10.3390/ijms151017411. Int J Mol Sci. 2014. PMID: 25268615 Free PMC article. Review. - Integrated multi-omics analyses reveal the TM4SF family genes with prognostic and therapeutic relevance in hepatocellular carcinoma.
Tang Q, Wang S, Li H, Liu J, Hu X, Zhao D, Di M. Tang Q, et al. Aging (Albany NY). 2024 Jan 10;16(1):593-616. doi: 10.18632/aging.205398. Epub 2024 Jan 10. Aging (Albany NY). 2024. PMID: 38206300 Free PMC article. - Regulation of DNA methylation machinery by epi-miRNAs in human cancer: emerging new targets in cancer therapy.
Karimzadeh MR, Pourdavoud P, Ehtesham N, Qadbeigi M, Asl MM, Alani B, Mosallaei M, Pakzad B. Karimzadeh MR, et al. Cancer Gene Ther. 2021 Apr;28(3-4):157-174. doi: 10.1038/s41417-020-00210-7. Epub 2020 Aug 10. Cancer Gene Ther. 2021. PMID: 32773776 Review. - Interaction of KLF6 and Sp1 regulates basigin-2 expression mediated proliferation, invasion and metastasis in hepatocellular carcinoma.
Kong LM, Yao L, Lu N, Dong YL, Zhang J, Wang YQ, Liu L, Zhang HL, Huang JG, Liao CG. Kong LM, et al. Oncotarget. 2016 May 10;7(19):27975-87. doi: 10.18632/oncotarget.8564. Oncotarget. 2016. PMID: 27057625 Free PMC article. - Emerging roles of ferroptosis in cardiovascular diseases.
Wang K, Chen XZ, Wang YH, Cheng XL, Zhao Y, Zhou LY, Wang K. Wang K, et al. Cell Death Discov. 2022 Sep 20;8(1):394. doi: 10.1038/s41420-022-01183-2. Cell Death Discov. 2022. PMID: 36127318 Free PMC article. Review.
References
- Tang ZY, Ye SL, Liu YK, et al. A decade’s studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:187–96. - PubMed
- Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–87. - PubMed
- Muramatsu T, Miyauchi T. Basigin (CD147): a multifunctional transmembrane protein involved in reproduction, neural function, inflammation and tumor invasion. Histol Histopathol. 2003;18:981–7. - PubMed
- Biswas C, Zhang Y, DeCastro R, et al. The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 1995;55:434–9. - PubMed
- Kanekura T, Miyauchi T, Tashiro M, et al. Basigin, a new member of the immunoglobulin superfamily: genes in different mammalian species, glycosylation changes in the molecule from adult organs and possible variation in the N-terminal sequences. Cell Struct Funct. 1991;16:23–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous