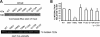Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration - PubMed (original) (raw)
. 2010 Sep 15;19(18):3591-8.
doi: 10.1093/hmg/ddq275. Epub 2010 Jul 14.
Affiliations
- PMID: 20631154
- PMCID: PMC2928130
- DOI: 10.1093/hmg/ddq275
Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration
Carlos A Murga-Zamalloa et al. Hum Mol Genet. 2010.
Abstract
Defects in biogenesis or function(s) of primary cilia are associated with numerous inherited disorders (called ciliopathies) that may include retinal degeneration phenotype. The cilia-expressed gene RPGR (retinitis pigmentosa GTPase regulator) is mutated in patients with X-linked retinitis pigmentosa (XLRP) and encodes multiple protein isoforms with a common N-terminal domain homologous to regulator of chromosome condensation 1 (RCC1), a guanine nucleotide exchange factor (GEF) for Ran GTPase. RPGR interacts with several ciliopathy proteins, such as RPGRIP1L and CEP290; however, its physiological role in cilia-associated functions has not been delineated. Here, we report that RPGR interacts with the small GTPase RAB8A, which participates in cilia biogenesis and maintenance. We show that RPGR primarily associates with the GDP-bound form of RAB8A and stimulates GDP/GTP nucleotide exchange. Disease-causing mutations in RPGR diminish its interaction with RAB8A and reduce the GEF activity. Depletion of RPGR in hTERT-RPE1 cells interferes with ciliary localization of RAB8A and results in shorter primary cilia. Our data suggest that RPGR modulates intracellular localization and function of RAB8A. We propose that perturbation of RPGR-RAB8A interaction, at least in part, underlies the pathogenesis of photoreceptor degeneration in XLRP caused by RPGR mutations.
Figures
Figure 1.
Interaction of RPGR with RAB8A. (A) Bovine retinal extract was prepared in the presence of the non-hydrolyzable GDP substrate and subjected to immunoprecipitation (IP) using anti-RAB8A, anti-RPGR antibodies or normal IgG and pre-immune (Pre-Imm) serum. Twenty percent of the protein used for IP was loaded in input lane. Vertical line represents different RPGR isoforms expressed in bovine retina, as described earlier (38,42). (B) A known-bait and known-prey yeast two-hybrid analysis. Upper panel represents the growth of yeast transformed with bait (RPGR1–15) and prey (RAB8A) constructs. Interaction between p53 (bait) and T antigen (T-Ag; prey) was used as a positive control, whereas laminin (LAM) and T-Ag association served as a negative control in this experiment. The lower panel represents β-galactosidase (blue) test of the growth assay shown in the upper panel. (C) Coomassie blue staining of the gel showing the purified WT GST-RPGR1–15 or GST proteins utilized in GST pulldown assay. Molecular mass markers are in kilo Daltons (kDa). (D and E) GST pulldown assay: 10 µg of purified GST-RPGR1–15 fusion protein or GST alone was incubated with 35S-labeled _in vitro_-translated RAB8A (C) or RAB11 (D) (or with indicated mutants). The proteins were collected by binding to Glutathione-Sepharose™ beads and analyzed by SDS–PAGE followed by autoradiography. Lanes are indicated. (F) Ciliated MDCK cells were stained with antibodies to RPGR (purple) and RAB8A (green). Immunostained cells were analyzed with OLYMPUS FV 500. White arrow in ‘Merge’ image shows co-localization of RPGR and RAB8A at the cilium. Nuclei (N) are stained with Hoechst (blue).
Figure 2.
Interaction of RPGR mutants with RAB8A. (A) To evaluate the interaction of RPGR mutants with RAB8A-T22N, we expressed and purified GST-RPGR1–15 WT and mutant proteins. Upper panel represents the Coomassie blue-stained gel showing that equivalent amount of individual protein used in the GST pulldown assay. The proteins were incubated with _in vitro_-translated 35S RAB8A-T22N, followed by pulldown using Glutathione-Sepharose beads. 35S RAB8A-T22N-positive signal was detected by autoradiography with phosphorimager (Storm 840 GE Healthcare) (lower panel). (B) Quantification of GST pulldown was analyzed from Coomassie blue-stained gels and the autoradiograms, using ImageJ software. Area and intensity of RAB8A signal was measured and plotted against the amount of the respective mutant protein used in the pulldown assay. Graph represents each ratio normalized against the ratio of the binding of WT-RPGR with 35S RAB8A-T22N. Error bars represent mean ± SD from five independent experiments. The level of significance was calculated with two-tailed _t_-test. *P < 0.001.
Figure 3.
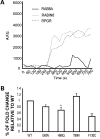
Modulation of RAB8A activity by RPGR. (A) Comparison of MANT-GTP incorporation into His-RAB8A as a function of time when incubated with RPGR or Rabin8. The activity of Rabin8 over RAB8A was used as positive control. AFU, arbitrary fluorescent units. (B) Fold change of RAB8A GDP/GTP exchange for each of the RPGR mutants relative to WT. Error bars represent mean ± SD from three independent experiments. Fold change was calculated as the ratio of RAB8A MANT-GTP incorporation after incubation with WT or mutant RPGR proteins with RAB8A MANT-GTP incorporation without incubation with RPGR (RAB8A intrinsic activity). Fold change by each mutant was compared with that depicted by WT RPGR from three independent experiments; level of significance was calculated with two-tailed _t_-test. *P < 0.05 and **P < 0.005.
Figure 4.
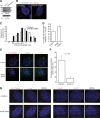
RPGR regulates ciliary localization of RAB8A. (A) Generation of RPGR-knockdown cells: lysates from stable transfected hTERT-RPE1 cells with control (scrambled) shRNA or RPGR shRNA were analyzed using anti-RPGR antibody or actin. Arrows represent distinct RPGR isoforms. (B) This panel represents cilia (red; acetylated α-tubulin) formation in control and RPGR-shRNA cells. Nuclei (blue) are stained with Hoechst. (C) Histograms show the distribution of cilia length in RPGR-shRNA-expressing cells. (D) There is increase in the number of cells with shorter cilia (1.5–3.9 µm) when treated with RPGR-shRNA compared with controls. Distribution of cilia length was calculated from three independent experiments with at least 150 cells analyzed in each experiment. Error bars represent standard deviation. (E) Knockdown of RPGR interferes with ciliary localization of RAB8A (green) to primary cilia (acetylated α-tubulin; red). Yellow in the Merge image indicates RAB8A localization at primary cilia. (F) Histogram showing decrease in number of cells with RAB8A at the cilium. We analyzed a total of 180 cells in each group and the experiment was repeated three times. (G) Knockdown of RPGR does not affect the localization of IFT88/Polaris (purple; left panel) to the cilium (red; Ac. α-tubulin; left panel) or of CEP290 (purple; right panel) to basal bodies (red; γ-tubulin; right panel).
Similar articles
- Multiprotein complexes of Retinitis Pigmentosa GTPase regulator (RPGR), a ciliary protein mutated in X-linked Retinitis Pigmentosa (XLRP).
Murga-Zamalloa C, Swaroop A, Khanna H. Murga-Zamalloa C, et al. Adv Exp Med Biol. 2010;664:105-14. doi: 10.1007/978-1-4419-1399-9_13. Adv Exp Med Biol. 2010. PMID: 20238008 Free PMC article. Review. - Prenylated retinal ciliopathy protein RPGR interacts with PDE6δ and regulates ciliary localization of Joubert syndrome-associated protein INPP5E.
Rao KN, Zhang W, Li L, Anand M, Khanna H. Rao KN, et al. Hum Mol Genet. 2016 Oct 15;25(20):4533-4545. doi: 10.1093/hmg/ddw281. Hum Mol Genet. 2016. PMID: 28172980 Free PMC article. - Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models.
Megaw R, Abu-Arafeh H, Jungnickel M, Mellough C, Gurniak C, Witke W, Zhang W, Khanna H, Mill P, Dhillon B, Wright AF, Lako M, Ffrench-Constant C. Megaw R, et al. Nat Commun. 2017 Aug 16;8(1):271. doi: 10.1038/s41467-017-00111-8. Nat Commun. 2017. PMID: 28814713 Free PMC article. - Interaction of ciliary disease protein retinitis pigmentosa GTPase regulator with nephronophthisis-associated proteins in mammalian retinas.
Murga-Zamalloa CA, Desai NJ, Hildebrandt F, Khanna H. Murga-Zamalloa CA, et al. Mol Vis. 2010 Jul 17;16:1373-81. Mol Vis. 2010. PMID: 20664800 Free PMC article. - More Than Meets the Eye: Current Understanding of RPGR Function.
Khanna H. Khanna H. Adv Exp Med Biol. 2018;1074:521-538. doi: 10.1007/978-3-319-75402-4_64. Adv Exp Med Biol. 2018. PMID: 29721984 Review.
Cited by
- OCRL localizes to the primary cilium: a new role for cilia in Lowe syndrome.
Luo N, West CC, Murga-Zamalloa CA, Sun L, Anderson RM, Wells CD, Weinreb RN, Travers JB, Khanna H, Sun Y. Luo N, et al. Hum Mol Genet. 2012 Aug 1;21(15):3333-44. doi: 10.1093/hmg/dds163. Epub 2012 Apr 27. Hum Mol Genet. 2012. PMID: 22543976 Free PMC article. - The New Era of Therapeutic Strategies for the Treatment of Retinitis Pigmentosa: A Narrative Review of Pathomolecular Mechanisms for the Development of Cell-Based Therapies.
Becherucci V, Bacci GM, Marziali E, Sodi A, Bambi F, Caputo R. Becherucci V, et al. Biomedicines. 2023 Sep 28;11(10):2656. doi: 10.3390/biomedicines11102656. Biomedicines. 2023. PMID: 37893030 Free PMC article. Review. - Accumulation of the Raf-1 kinase inhibitory protein (Rkip) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies.
Murga-Zamalloa CA, Ghosh AK, Patil SB, Reed NA, Chan LS, Davuluri S, Peränen J, Hurd TW, Rachel RA, Khanna H. Murga-Zamalloa CA, et al. J Biol Chem. 2011 Aug 12;286(32):28276-86. doi: 10.1074/jbc.M111.237560. Epub 2011 Jun 17. J Biol Chem. 2011. PMID: 21685394 Free PMC article. - Loss of human disease protein retinitis pigmentosa GTPase regulator (RPGR) differentially affects rod or cone-enriched retina.
Rao KN, Li L, Zhang W, Brush RS, Rajala RV, Khanna H. Rao KN, et al. Hum Mol Genet. 2016 Apr 1;25(7):1345-56. doi: 10.1093/hmg/ddw017. Epub 2016 Jan 24. Hum Mol Genet. 2016. PMID: 26908598 Free PMC article. - Rpgrip1 is required for rod outer segment development and ciliary protein trafficking in zebrafish.
Raghupathy RK, Zhang X, Liu F, Alhasani RH, Biswas L, Akhtar S, Pan L, Moens CB, Li W, Liu M, Kennedy BN, Shu X. Raghupathy RK, et al. Sci Rep. 2017 Dec 4;7(1):16881. doi: 10.1038/s41598-017-12838-x. Sci Rep. 2017. PMID: 29203866 Free PMC article.
References
- Scholey J.M., Anderson K.V. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi:10.1016/j.cell.2006.04.013. - DOI - PubMed
- Singla V., Reiter J.F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi:10.1126/science.1124534. - DOI - PubMed
- Dawe H.R., Farr H., Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell. Sci. 2007;120:7–15. doi:10.1242/jcs.03305. - DOI - PubMed
- Gerdes J.M., Davis E.E., Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi:10.1016/j.cell.2009.03.023. - DOI - PMC - PubMed
- Lancaster M.A., Gleeson J.G. The primary cilium as a cellular signaling center: lessons from disease. Curr. Opin. Genet. Dev. 2009;19:220–229. doi:10.1016/j.gde.2009.04.008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous