Viruses in the faecal microbiota of monozygotic twins and their mothers - PubMed (original) (raw)
Comparative Study
. 2010 Jul 15;466(7304):334-8.
doi: 10.1038/nature09199.
Affiliations
- PMID: 20631792
- PMCID: PMC2919852
- DOI: 10.1038/nature09199
Comparative Study
Viruses in the faecal microbiota of monozygotic twins and their mothers
Alejandro Reyes et al. Nature. 2010.
Abstract
Viral diversity and life cycles are poorly understood in the human gut and other body habitats. Phages and their encoded functions may provide informative signatures of a human microbiota and of microbial community responses to various disturbances, and may indicate whether community health or dysfunction is manifest after apparent recovery from a disease or therapeutic intervention. Here we report sequencing of the viromes (metagenomes) of virus-like particles isolated from faecal samples collected from healthy adult female monozygotic twins and their mothers at three time points over a one-year period. We compared these data sets with data sets of sequenced bacterial 16S ribosomal RNA genes and total-faecal-community DNA. Co-twins and their mothers share a significantly greater degree of similarity in their faecal bacterial communities than do unrelated individuals. In contrast, viromes are unique to individuals regardless of their degree of genetic relatedness. Despite remarkable interpersonal variations in viromes and their encoded functions, intrapersonal diversity is very low, with >95% of virotypes retained over the period surveyed, and with viromes dominated by a few temperate phages that exhibit remarkable genetic stability. These results indicate that a predatory viral-microbial dynamic, manifest in a number of other characterized environmental ecosystems, is notably absent in the very distal intestine.
Figures
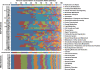
Fig. 1. Classification of viruses present in VLP preparations generated from fecal samples collected from four families of MZ twins and their mothers
Prophages are classified based on their bacterial host taxonomy. Prominent bacterial phyla are represented by different colors (Proteobacteria, blue; Firmicutes green; Bacteroidetes, red; Actinobacteria, black). Class-level taxa within these phyla are indicated. Phage and eukaryotic viruses are sorted according to taxonomy. Nomenclature used for VLP preparations from fecal biospecimens: F, family; T1, co-twin 1; T2, co-twin 2; M, mother of co-twins. Time points (1–3), and technical replicates (R) produced from a given sample are noted. The color bar at the bottom of the figure provides a reference key for the percent coverage of a viral genome in the NR_Viral_DB by reads from given VLP virome dataset (data normalized using 14,000 randomly selected reads/dataset).
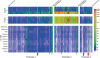
Fig. 2. A sample-by-sample view of the proportional representation of KEGG second level pathways in sequenced VLP-associated viromes and gut microbiomes
Known or predicted proteins encoded by viruses in the NR_Viral_DB, fecal VLP-derived viromes, 121 sequenced reference human gut-associated microbial genomes, and fecal microbiomes are shown. See Fig. 1 for sample nomenclature.
Fig. 3. Gnotobiotic mice reveal in vivo activation of the transcriptome of a Marvinbryantia formatexigens prophage
Shown are the three predicted prophages present in M. formatexigens and the levels of expression of their ORFs in cecal and fecal micobial communities harvested from gnotobiotic mice co-colonized with Bacteroides thetaiotaomicron. Expression levels for genes in each prophage genome are from normalized RNA-Seq read count data (see color key; normalization based on sequencing effort and length of each predicted ORF). Active expression is defined as a normalized read count >100. R, technical replicate shows the reproducibility of the method for performing RNA-Seq analysis. RNA-Seq data are also presented for each prophage genome in M. formatexigens during mid-log phase growth in defined medium containing different carbon sources (NAG, N-acetylglucosamine). Red arrows indicate the position of toxin/anti-toxin gene pairs. Green arrows denote genes with hypothetical functions that are expressed in more than 50% of the conditions tested. ORF designations for the first and last genes in the predicted genomes of each prophage are provided.
Similar articles
- Census of the viral metagenome within an activated sludge microbial assemblage.
Parsley LC, Consuegra EJ, Thomas SJ, Bhavsar J, Land AM, Bhuiyan NN, Mazher MA, Waters RJ, Wommack KE, Harper WF Jr, Liles MR. Parsley LC, et al. Appl Environ Microbiol. 2010 Apr;76(8):2673-7. doi: 10.1128/AEM.02520-09. Epub 2010 Feb 12. Appl Environ Microbiol. 2010. PMID: 20154108 Free PMC article. - Virome Diversity Correlates with Intestinal Microbiome Diversity in Adult Monozygotic Twins.
Moreno-Gallego JL, Chou SP, Di Rienzi SC, Goodrich JK, Spector TD, Bell JT, Youngblut ND, Hewson I, Reyes A, Ley RE. Moreno-Gallego JL, et al. Cell Host Microbe. 2019 Feb 13;25(2):261-272.e5. doi: 10.1016/j.chom.2019.01.019. Cell Host Microbe. 2019. PMID: 30763537 Free PMC article. - The gut virome of the protochordate model organism, Ciona intestinalis subtype A.
Leigh BA, Djurhuus A, Breitbart M, Dishaw LJ. Leigh BA, et al. Virus Res. 2018 Jan 15;244:137-146. doi: 10.1016/j.virusres.2017.11.015. Epub 2017 Nov 15. Virus Res. 2018. PMID: 29155033 - Metaviromics coupled with phage-host identification to open the viral 'black box'.
Moon K, Cho JC. Moon K, et al. J Microbiol. 2021 Mar;59(3):311-323. doi: 10.1007/s12275-021-1016-9. Epub 2021 Feb 23. J Microbiol. 2021. PMID: 33624268 Review. - Diversity and Ecology of Viruses in Hyperarid Desert Soils.
Zablocki O, Adriaenssens EM, Cowan D. Zablocki O, et al. Appl Environ Microbiol. 2015 Nov 20;82(3):770-7. doi: 10.1128/AEM.02651-15. Print 2016 Feb 1. Appl Environ Microbiol. 2015. PMID: 26590289 Free PMC article. Review.
Cited by
- High throughput sequencing provides exact genomic locations of inducible prophages and accurate phage-to-host ratios in gut microbial strains.
Zünd M, Ruscheweyh HJ, Field CM, Meyer N, Cuenca M, Hoces D, Hardt WD, Sunagawa S. Zünd M, et al. Microbiome. 2021 Mar 29;9(1):77. doi: 10.1186/s40168-021-01033-w. Microbiome. 2021. PMID: 33781335 Free PMC article. - A test for the "physiological phagemia" hypothesis-natural intestinal coliphages do not penetrate to the blood in horses.
Letarova M, Strelkova D, Nevolina S, Letarov A. Letarova M, et al. Folia Microbiol (Praha). 2012 Jan;57(1):81-3. doi: 10.1007/s12223-011-0096-z. Epub 2012 Jan 11. Folia Microbiol (Praha). 2012. PMID: 22234557 No abstract available. - Antibiotics in feed induce prophages in swine fecal microbiomes.
Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D, Stanton TB. Allen HK, et al. mBio. 2011 Nov 29;2(6):e00260-11. doi: 10.1128/mBio.00260-11. Print 2011. mBio. 2011. PMID: 22128350 Free PMC article. - Phage therapy: Targeting intestinal bacterial microbiota for the treatment of liver diseases.
Fujiki J, Schnabl B. Fujiki J, et al. JHEP Rep. 2023 Sep 23;5(12):100909. doi: 10.1016/j.jhepr.2023.100909. eCollection 2023 Dec. JHEP Rep. 2023. PMID: 37965159 Free PMC article. Review. - Metagenomic Analysis of the Respiratory Microbiome of a Broiler Flock from Hatching to Processing.
Mulholland KA, Robinson MG, Keeler SJ, Johnson TJ, Weber BW, Keeler CL Jr. Mulholland KA, et al. Microorganisms. 2021 Mar 31;9(4):721. doi: 10.3390/microorganisms9040721. Microorganisms. 2021. PMID: 33807233 Free PMC article.
References
- Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. - PubMed
- Angly F, et al. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol. 2009;11:2863–2873. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases