Confining euchromatin/heterochromatin territory: jumonji crosses the line - PubMed (original) (raw)
Review
Confining euchromatin/heterochromatin territory: jumonji crosses the line
Hisashi Tamaru. Genes Dev. 2010.
Abstract
Heterochromatin is typically highly condensed, gene-poor, and transcriptionally silent, whereas euchromatin is less condensed, gene-rich, and more accessible to transcription. Besides acting as a graveyard for selfish mobile DNA repeats, heterochromatin contributes to important biological functions, such as chromosome segregation during cell division. Multiple features of heterochromatin-including the presence or absence of specific histone modifications, DNA methylation, and small RNAs-have been implicated in distinguishing heterochromatin from euchromatin in various organisms. Cells malfunction if the genome fails to restrict repressive chromatin marks within heterochromatin domains. How euchromatin and heterochromatin territories are confined remains poorly understood. Recent studies from the fission yeast Schizosaccharomyces pombe, the flowering plant Arabidopsis thaliana, and the filamentous fungus Neurospora crassa have revealed a new role for Jumonji C (JmjC) domain-containing proteins in protecting euchromatin from heterochromatin marks.
Figures
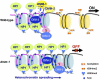
Figure 1.
Model for the involvement of DMM-1 in preventing spreading of heterochromatin into euchromatin. In N. crassa, chromatin regions containing A:T-rich DNA sequences (pink fiber) that have been subjected to RIP constitute heterochromatin (nucleosomes in blue). RNAi components are dispensable for heterochromatin targeting. It is thought that an unidentified protein (X) that recognizes TpA/ApT base pairs recruits DIM-5 HMT to establish H3K9me. HP1 forms a complex with the DMT DIM-2 and binds to H3K9me via its chromodomain, allowing DIM-2 to methylate nucleosomal DNA (5mC) in all sequence contexts. HP1 also recruits DMM-1 and its associated protein, DMM-2, to RIP'd heterochromatic regions. The HP1/DMM-1/DMM-2 complex is preferentially localized to edges of heterochromatin to block HP1/DIM-2 and/or DIM-5 activity. DMM-1/DMM-2 may bind to both RIP'd and non-RIP'd sites of heterochromatin/euchromatin boundaries through the DMM-1 cysteine-rich domains that are implicated in binding to an unidentified histone mark (question mark) and the DMM-2 Zn(II)2Cys6 DNA-binding motif, respectively. A dmm-1 mutation allows HP1/DIM-2 to spread across non-RIP'd euchromatic regions (nucleosomes in yellow) and methylate non-RIP'd DNA (black fiber). In this situation, 5mC attracts DIM-5 to methylate H3K9, presumably via an unidentified DNA methyl-binding protein (Y). Heterochromatin spreading can silence nearby genes. Although the JmjC domain of DMM-1 is required for its function, histone lysine demethylase (KDM) activity has not been detected for DMM-1 (Honda et al. 2010).
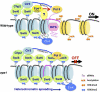
Figure 2.
Model for the involvement of Epe1 in preventing spreading of heterochromatin into euchromatin. In S. pombe, the IRC elements (pink fiber) located at the boundaries of heterochromatin are transcribed by Pol II within the context of heterochromatin, are processed into siRNAs by the RITS complex, and serve as heterochromatin barrier elements. A DNA-binding factor or the RNAi pathway targets Clr4 HMT to heterochromatic regions (nucleosomes in blue) for H3K9me, which provides a binding site for the chromodomain of Swi6 (homolog of HP1). Swi6 recruits the JmjC domain protein Epe1. The histone deacetylase Clr3 is recruited by Swi6 and another chromodomain protein, Chp2. Clr3 limits Pol II accessibility to heterochromatic repeats and the IRC barrier elements, whereas Epe1 promotes its transcription. There is evidence for an unidentified protein (questionmark) that specifically targets Epe1 to heterochromatin/euchromatin boundaries. High concentrations of Epe1 and the resulting high levels of transcription at _IRC_s presumably provide an “open” chromatin environment that counteracts the repressive effects of heterochromatin. The balance between the opposing activities of Clr3 and Epe1 is critical for confining heterochromatin and its associated factors within a defined territory (Zofall and Grewal 2006). An epe1 mutation leads predominantly to spreading of heterochromatin across euchromatic regions (nucleosomes in yellow) through self-reinforcing reactions between H3K9me and Swi6. Binding of the Clr4 chromodomain to H3K9me facilitates methylation of adjacent nucleosomes. Heterochromatin spreading can cause silencing of neighboring genes. It may be envisioned that, in a small population of cells, loss of Epe1 completely shuts off transcription of heterochromatic repeats and subsequent processing into siRNAs, which are essential for the stable maintenance of heterochromatin, thereby resulting in heterochromatin contraction. Histone lysine demethylase (KDM) activity has not been detected with Epe1 (Zofall and Grewal 2006).
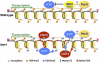
Figure 3.
Model for the involvement of IBM1 in protecting transcribed genes from CHG methylation. In A. thaliana, DNA 5mC of genes is limited to CG sites. IBM1 prevents KYP HMT from inappropriate deposition of H3K9me on the bodies of active genes by either suppressing KYP activity or demethylating H3K9me in a transcription-coupled manner. It is not known whether IBM and KYP interact with Pol II. An ibm1 mutation allows KYP to methylate H3K9 within transcribed regions. The chromodomain of the DMT CMT3 recognizes dual methylation marks at H3K9 and H3K27, and methylates DNA preferentially in the CHG context. The SRA domain of KYP binds to 5mC to facilitate methylation of adjacent nucleosomes. The ectopic CHG methylation of the gene body is not invariably associated with transcriptional repression. The central portions of long transcribed genes are most frequently subjected to _ibm1_-induced de novo methylation by unknown mechanisms. RdDM components do not seem to be involved in the process. The JmjC domain of IBM1 has all of the key amino acid residues critical for lysine demethylase (KDM) activity, but the activity has not been detected with IBM1 (Saze et al. 2008; Miura et al. 2009). (P) Promoter; (T) terminator.
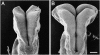
Figure 4.
Abnormal groove formation on the neural plate of jumonji homozygous mutant mouse embryos. Scanning electron micrographs of dorsal views of wild-type (A) and jumonji mutant (B) mouse embryos at embryonic day 8.5 (nine-somite stage). Bar, 100 μm. Photographs courtesy of Takashi Takeuchi.
Similar articles
- Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin.
Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Li F, et al. Cell. 2008 Oct 17;135(2):272-83. doi: 10.1016/j.cell.2008.08.036. Cell. 2008. PMID: 18957202 Free PMC article. - The genome organization of Neurospora crassa at high resolution uncovers principles of fungal chromosome topology.
Rodriguez S, Ward A, Reckard AT, Shtanko Y, Hull-Crew C, Klocko AD. Rodriguez S, et al. G3 (Bethesda). 2022 May 6;12(5):jkac053. doi: 10.1093/g3journal/jkac053. G3 (Bethesda). 2022. PMID: 35244156 Free PMC article. - The JmjC domain protein Epe1 prevents unregulated assembly and disassembly of heterochromatin.
Trewick SC, Minc E, Antonelli R, Urano T, Allshire RC. Trewick SC, et al. EMBO J. 2007 Nov 14;26(22):4670-82. doi: 10.1038/sj.emboj.7601892. Epub 2007 Oct 18. EMBO J. 2007. PMID: 17948055 Free PMC article. - Histone Methylation by SET Domain Proteins in Fungi.
Freitag M. Freitag M. Annu Rev Microbiol. 2017 Sep 8;71:413-439. doi: 10.1146/annurev-micro-102215-095757. Epub 2017 Jul 17. Annu Rev Microbiol. 2017. PMID: 28715960 Review. - Molecular Complexes at Euchromatin, Heterochromatin and Centromeric Chromatin.
Morrison O, Thakur J. Morrison O, et al. Int J Mol Sci. 2021 Jun 28;22(13):6922. doi: 10.3390/ijms22136922. Int J Mol Sci. 2021. PMID: 34203193 Free PMC article. Review.
Cited by
- RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats.
Lee TF, Gurazada SG, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC. Lee TF, et al. Epigenetics. 2012 Jul;7(7):781-95. doi: 10.4161/epi.20290. Epub 2012 Jul 1. Epigenetics. 2012. PMID: 22647529 Free PMC article. - Understanding the DNA damage response in order to achieve desired gene editing outcomes in mosquitoes.
Overcash JM, Aryan A, Myles KM, Adelman ZN. Overcash JM, et al. Chromosome Res. 2015 Feb;23(1):31-42. doi: 10.1007/s10577-014-9450-8. Chromosome Res. 2015. PMID: 25596822 Free PMC article. Review. - Accumulation of DNA damage-induced chromatin alterations in tissue-specific stem cells: the driving force of aging?
Schuler N, Rübe CE. Schuler N, et al. PLoS One. 2013 May 17;8(5):e63932. doi: 10.1371/journal.pone.0063932. Print 2013. PLoS One. 2013. PMID: 23691119 Free PMC article. - ROS, Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases.
Davalli P, Mitic T, Caporali A, Lauriola A, D'Arca D. Davalli P, et al. Oxid Med Cell Longev. 2016;2016:3565127. doi: 10.1155/2016/3565127. Epub 2016 May 10. Oxid Med Cell Longev. 2016. PMID: 27247702 Free PMC article. Review. - Histone and Histone Acetylation-Related Alterations of Gene Expression in Uninvolved Psoriatic Skin and Their Effects on Cell Proliferation, Differentiation, and Immune Responses.
Romhányi D, Szabó K, Kemény L, Groma G. Romhányi D, et al. Int J Mol Sci. 2023 Sep 26;24(19):14551. doi: 10.3390/ijms241914551. Int J Mol Sci. 2023. PMID: 37833997 Free PMC article.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938 - PMC - PubMed
- Baker WK 1968. Position-effect variegation. Adv Genet 14: 133–169 - PubMed
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources