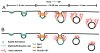Actin dynamics and endocytosis in yeast and mammals - PubMed (original) (raw)
Review
Actin dynamics and endocytosis in yeast and mammals
Brian J Galletta et al. Curr Opin Biotechnol. 2010 Oct.
Abstract
Tight regulation of the actin cytoskeleton is critical for many cell functions, including various forms of cellular uptake. Clathrin-mediated endocytosis (CME) is one of the main methods of uptake in many cell types. An intact and properly regulated actin cytoskeleton is required for CME in Saccharomyces cerevisiae. Yeast CME requires the proper regulation of actin polymerization, filament cross-linking, and filament disassembly. Recent studies also point to a role for F-BAR and BAR-domain containing proteins in linking the processes of generating and sensing plasma membrane curvature with those regulating the actin cytoskeleton. Many of these same proteins are conserved in mammalian CME. However, until recently the requirement for actin in mammalian CME was less clear. Several recent studies in mammalian cells provide new support for an actin requirement in the invagination and late stages of CME. This review focuses on the regulation of the actin cytoskeleton during CME in yeast and the emerging evidence for a role for actin during mammalian CME.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
Figure 1
Major models for actin assembly during endocytosis derived from the results of numerous works described and referenced herein. In both A and B the initial curvature of the membrane is generated by clathrin and additional endocytic proteins, such as F-BAR containing Syp1. Syp1 can inhibit WASp/Las17-Arp2/3 mediated actin assembly and may serve to keep actin polymerization inhibited during early steps of CME. As invagination proceeds, the changing membrane curvature may be sensed by other BAR domain containing proteins, for example Bzz1, which is recruited along with other endocytic proteins and regulators of Arp2/3. In model A, a ring of NPFs activate Arp2/3 which nucleates an actin network that flows away from the plasma membrane. Proteins of the endocytic coat link the membrane to this flowing network and this provides the force to invaginate further into the cell. The amphiphysin proteins, Rvs161 and Rvs167, drive membrane scission. In model B, actin is nucleated along the sides of the membrane tubule, perhaps in response to the loss of Syp1, and the recruitment of Bzz1, which can promote actin nucleation. The growing barbed ends of the actin push on the membrane tubule, squeezing it, driving elongation and assisting the amphiphysin proteins during membrane scission. In this model, the actin network is in a position to drive movement after the vesicle has been freed from the membrane. The best models for how actin polymerization is utilized during clathrin-mediated endocytosis in mammals are similar to the model presented in B [64, 65]
Similar articles
- Roles for actin assembly in endocytosis.
Mooren OL, Galletta BJ, Cooper JA. Mooren OL, et al. Annu Rev Biochem. 2012;81:661-86. doi: 10.1146/annurev-biochem-060910-094416. Annu Rev Biochem. 2012. PMID: 22663081 Review. - The mammalian endocytic cytoskeleton.
Abouelezz A, Almeida-Souza L. Abouelezz A, et al. Eur J Cell Biol. 2022 Apr;101(2):151222. doi: 10.1016/j.ejcb.2022.151222. Epub 2022 Apr 3. Eur J Cell Biol. 2022. PMID: 35413660 Review. - Bending over backwards: BAR proteins and the actin cytoskeleton in mammalian receptor-mediated endocytosis.
Armstrong G, Olson MF. Armstrong G, et al. Eur J Cell Biol. 2022 Jun-Aug;101(3):151257. doi: 10.1016/j.ejcb.2022.151257. Epub 2022 Jul 16. Eur J Cell Biol. 2022. PMID: 35863103 Review. - A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis.
Yarar D, Waterman-Storer CM, Schmid SL. Yarar D, et al. Mol Biol Cell. 2005 Feb;16(2):964-75. doi: 10.1091/mbc.e04-09-0774. Epub 2004 Dec 15. Mol Biol Cell. 2005. PMID: 15601897 Free PMC article. - Actin and endocytosis in budding yeast.
Goode BL, Eskin JA, Wendland B. Goode BL, et al. Genetics. 2015 Feb;199(2):315-58. doi: 10.1534/genetics.112.145540. Genetics. 2015. PMID: 25657349 Free PMC article. Review.
Cited by
- Depolymerization of cortical actin inhibits UT-A1 urea transporter endocytosis but promotes forskolin-stimulated membrane trafficking.
Xu G, Su H, Carter CB, Fröhlich O, Chen G. Xu G, et al. Am J Physiol Cell Physiol. 2012 Apr 1;302(7):C1012-8. doi: 10.1152/ajpcell.00440.2011. Epub 2012 Jan 18. Am J Physiol Cell Physiol. 2012. PMID: 22262062 Free PMC article. - Genetic and epigenetic changes in chromosomally stable and unstable progeny of irradiated cells.
Baulch JE, Aypar U, Waters KM, Yang AJ, Morgan WF. Baulch JE, et al. PLoS One. 2014 Sep 24;9(9):e107722. doi: 10.1371/journal.pone.0107722. eCollection 2014. PLoS One. 2014. PMID: 25251398 Free PMC article. - A new role for myosin II in vesicle fission.
Flores JA, Balseiro-Gomez S, Cabeza JM, Acosta J, Ramirez-Ponce P, Ales E. Flores JA, et al. PLoS One. 2014 Jun 24;9(6):e100757. doi: 10.1371/journal.pone.0100757. eCollection 2014. PLoS One. 2014. PMID: 24959909 Free PMC article. - Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis.
Collins A, Warrington A, Taylor KA, Svitkina T. Collins A, et al. Curr Biol. 2011 Jul 26;21(14):1167-75. doi: 10.1016/j.cub.2011.05.048. Epub 2011 Jun 30. Curr Biol. 2011. PMID: 21723126 Free PMC article. - Combinatorial genetic analysis of a network of actin disassembly-promoting factors.
Ydenberg CA, Johnston A, Weinstein J, Bellavance D, Jansen S, Goode BL. Ydenberg CA, et al. Cytoskeleton (Hoboken). 2015 Jul;72(7):349-61. doi: 10.1002/cm.21231. Epub 2015 Aug 22. Cytoskeleton (Hoboken). 2015. PMID: 26147656 Free PMC article.
References
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. - PubMed
- Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 GM083538/GM/NIGMS NIH HHS/United States
- R01 GM038542/GM/NIGMS NIH HHS/United States
- F32 GM 083538/GM/NIGMS NIH HHS/United States
- GM 38542/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases