Type I interferons regulate cytolytic activity of memory CD8(+) T cells in the lung airways during respiratory virus challenge - PubMed (original) (raw)
Type I interferons regulate cytolytic activity of memory CD8(+) T cells in the lung airways during respiratory virus challenge
Jacob E Kohlmeier et al. Immunity. 2010.
Abstract
Memory CD8(+) T cells in the lung airways provide protection from secondary respiratory virus challenge by limiting early viral replication. Here, we demonstrate that although airway-resident memory CD8(+) T cells were poorly cytolytic, memory CD8(+) T cells recruited to the airways early during a recall response showed markedly enhanced cytolytic ability. This enhanced lytic activity did not require cognate antigen stimulation, but rather was dependent on STAT1 transcription factor signaling through the interferon-alpha receptor (Ifnar1), resulting in the antigen-independent expression of granzyme B protein in both murine and human virus-specific T cells. Signaling through Ifnar1 was required for the enhanced lytic activity and control of early viral replication by memory CD8(+) T cells in the lung airways. These findings demonstrate that innate inflammatory signals act directly on memory T cells, enabling them to rapidly destroy infected host cells once they enter infected tissues.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
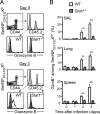
Figure 1. Memory CD8+ T cells recruited to the airways after virus infection have enhanced lytic activity
C57Bl/6 mice were infected with Sendai virus and rested for 45 days. Mice were then challenged with PBS or x31 influenza virus, and CD8+ T cells from the BAL were purified by cell sorting 3 days later. (A) Sorted CD8+ T cells from the BAL were incubated with congenic (CD45.1+) target cells pulsed with SenNP324-332 (CFSE lo) or γHV p79524-531 (CFSE hi) for 5 hours. Live target cells (CD45.1+ and Propidium iodide-) were then analyzed by flow cytometry and the ratio of SenNP targets to γHV p79 targets was used to calculate specific lysis. Representative flow cytometry plots for each treatment are shown. (B) The percent specific lysis of SenNP-pulsed targets after incubation with CD8+ BAL T cells from PBS-treated (open circles) or x31-infected (closed squares) mice at various effector to target (E:T) ratios. The data are representative of 5 independent experiments with 5-8 replicate wells per group.
Figure 2. Granzyme B protein expression is up-regulated in memory CD8+ T cells in response to inflammation
C57Bl/6 mice were infected with Sendai virus and rested for 45 days prior to challenge with x31 influenza virus. (A) Representative staining of isotype (shaded histograms) or granzyme B (open histograms) antibodies in the BAL, lung, and spleen on days 0-5 post-x31 challenge. All plots are gated on SenNP324-332Kb+ cells. (B) The frequency of gzmB+ cells among total SenNP324-332Kb+ cells in the BAL, lung, and spleen. Each symbol represents an individual mouse and significance was determined by comparing each timepoint with the values from day 0. (C) The frequency of of gzmB+ cells among total SenNP324-332Kb+ cells in the BAL after intranasal administration of Poly I:C (open bars) or CpG (closed bars). (D) Sorted CD8+ T cells from the BAL of Sendai memory mice on day 3 after influenza challenge were incubated with congenic targets at a 4:1 E:T ratio with the indicated inhibitors of cytotoxicity pathways. Each symbol represents the percent specific lysis of a replicate well. The data represent 4 independent experiments with at least 5 mice per group (A and B), 3 independent experiments with 4 mice per group (C), or 2 independent experiments with 5 wells per treatment (D).
Figure 3. Granzyme B protein expression and memory T cell lytic activity is limited to cells recently recruited to the airways
(A) C57Bl/6 mice were infected with Sendai virus and rested for 45 days prior to challenge with x31 influenza virus. Plots are gated on SenNP324-332Kb+ cells and show the expression of gzmB in airway resident (CD11alo) and recently recruited (CD11ahi) Sendai memory CD8+ T cells on days 0-5 after influenza x31 challenge. Quadrant gates were set based on isotype staining. (B) The frequency of airway-resident (open bars) and recently-recruited (shaded bars) Sendai-specific cells is graphed as the mean ± s.d. for days 0-5 after influenza challenge. (C) CD8+ T cells from the BAL of Sendai memory mice on day 3 after influenza challenge were sorted into airway-resident and recently-recruited populations based on CD11a expression. The percent specific lysis of SenNP-pulsed targets is graphed as the mean ± s.d. for each population at the indicated E:T ratios. The data are representative of 3 independent experiments with 4 mice per group (A and B) or 5 replicate wells per group (C).
Figure 4. STAT1 is required for the antigen-independent expression of gzmB protein in memory CD8+ T cells
Mixed BM chimeras were generated from WT congenic (CD45.2-) and _Stat1_-/- donors, infected with Sendai virus, and rested for 45 days prior to influenza x31 challenge. (A) Representative staining of gzmB protein in WT and _Stat1_-/- Sendai-specific memory CD8+ T cells in the lung prior to (day 0, top panels) or on day 3 (bottom panels) after influenza challenge. The shaded histograms are isotype staining, whereas the open histograms are gzmB staining. (B) The frequency of WT (open bars) and _Stat1_-/- (shaded bars) Sendai-specific memory CD8+ T cells expressing gzmB at the indicated timepoints is graphed as the mean ± s.d. (* p<0.05; ** p<0.005). The data are representative of 3 independent experiments with 3-5 mice per timepoint.
Figure 5. Signaling through Ifnar1 is necessary for antigen-independent gzmB protein expression and lytic activity of memory CD8+ T cells in the lung airways
Mixed BM chimeras were generated from WT and cytokine receptor-deficient donors (_Ifnar1_-/-, _Ifngr1_-/-, and _Il27ra_-/-), infected Sendai virus, and rested 45-60 days prior to influenza x31 challenge. (A) Representative isotype (shaded histgrams) and gzmB (open histograms) staining of cytokine receptor-deficient (upper panels) and WT (lower panels) Sendai-specific memory CD8+ T cells in the airways from each group of chimeras at day 3 post-x31 challenge. Histograms are gated on SenNP324-332Kb+ cells. (B-D) The frequency of WT (open bars) and cytokine receptor-deficient (shaded bars) gzmB+ Sendai-specific memory T cells in various tissues prior to or on day 3 following x31 challenge for (B) _Ifnar1_-/-, (C) _Ifngr1_-/-, and (D) _Il27ra_--/- mixed BM chimeras (* p<0.01). (E) Phenotypic analysis of WT (CD90.1+, black) and _Ifnar1_-/- (CD90.1-, grey) Sendai-specific memory CD8+ T cells in the airways of mixed BM chimeras on day 3 after x31 challenge. (F) WT and _Ifnar1_-/- CD8+ T cells were sorted from the airways of mixed BM chimera Sendai memory mice on day 3 after x31 challenge. The percent specific lysis of SenNP-pulsed targets following a 5 hour incubation with sorted WT (open circles) or _Ifnar1_-/- (shaded circles) CD8+ T cells is graphed as the mean ± s.d. for each population at the indicated E:T ratios. The data are representative of 3 independent experiments with at least 5 mice per timepoint.
Figure 6. Type I interferons are sufficient to induce gzmB protein expression in both murine and human virus-specific memory CD8+ T cells
(A) C57BL/6 mice were infected with Sendai virus and rested for 45 days. Splenocytes from these mice were incubated in vitro with or without IFN-α or IFN-β (2×104 Units / ml) for 36 hours and gzmB protein expression was measured by flow cytometry. Representative staining is shown for isotype (shaded histogram) or anti-gzmB (open histogram) gated on SenNP324-332Kb+ cells. (B) The percent change in gzmB expression was graphed for IFN-α or IFN-β treatment compared to control samples. Each symbol represents an individual mouse and statistical significance was determined using a paired t-test. (C) Representative staining and gating strategy to identify influenza-specific memory CD8+ T cells from the peripheral blood of healthy donors. (D) The frequency of A2 FluM1-specific cells among total CD8+ T cells versus the frequency of gzmB+ FluM1-specific cells is graphed for each individual donor prior to in vitro stimulation. (E) Representative staining of gzmB protein in A2 FluM1+ cells following in vitro culture with or without IFN-α or IFN-β (2×104 Units / ml). (F) The percent change in gzmB expression was graphed for IFN-α or IFN-β treatment compared to control samples. Each symbol represents an individual donor and statistical significance was determined using a paired t-test. The data are representative of 3 (A,B) or 2 (C-F) independent experiments.
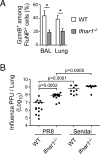
Figure 7. Ifnar1 signaling is required for maximal gzmB protein expression and T cell-mediated protection when cognate antigen is present
WT × _Ifnar1_-/- mixed BM chimeras were infected with PR8 influenza and rested 45 days prior to challenge with x31 influenza. (A) The frequency of gzmB+ cells among WT (open bars) or _Ifnar1_-/- (shaded bars) FluNP-specific T cells is graphed as the mean ± s.d. (*p<0.02). (B) Complete BM chimeras were generated with BM from either WT (C57BL/6) or _Ifnar1_-/- donors (BM was not mixed), infected with influenza PR8 or Sendai virus, and rested for 60 days. All mice were challenged with influenza x31 and viral titers were measured on day 3. Each symbol represents an individual mouse. The data are representative of 3 (A, 5 mice per timepoint) or 2 (B) independent experiments.
Comment in
- Forewarned is forearmed.
La Gruta N, Turner SJ. La Gruta N, et al. Immunity. 2010 Jul 23;33(1):5-6. doi: 10.1016/j.immuni.2010.07.007. Immunity. 2010. PMID: 20643333
Similar articles
- Nonspecific recruitment of memory CD8+ T cells to the lung airways during respiratory virus infections.
Ely KH, Cauley LS, Roberts AD, Brennan JW, Cookenham T, Woodland DL. Ely KH, et al. J Immunol. 2003 Feb 1;170(3):1423-9. doi: 10.4049/jimmunol.170.3.1423. J Immunol. 2003. PMID: 12538703 - Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections.
Hogan RJ, Usherwood EJ, Zhong W, Roberts AA, Dutton RW, Harmsen AG, Woodland DL. Hogan RJ, et al. J Immunol. 2001 Feb 1;166(3):1813-22. doi: 10.4049/jimmunol.166.3.1813. J Immunol. 2001. PMID: 11160228 - Airway-Resident Memory CD8 T Cells Provide Antigen-Specific Protection against Respiratory Virus Challenge through Rapid IFN-γ Production.
McMaster SR, Wilson JJ, Wang H, Kohlmeier JE. McMaster SR, et al. J Immunol. 2015 Jul 1;195(1):203-9. doi: 10.4049/jimmunol.1402975. Epub 2015 May 29. J Immunol. 2015. PMID: 26026054 Free PMC article. - Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus.
Slütter B, Pewe LL, Kaech SM, Harty JT. Slütter B, et al. Immunity. 2013 Nov 14;39(5):939-48. doi: 10.1016/j.immuni.2013.09.013. Immunity. 2013. PMID: 24238342 Free PMC article. - Type I interferon plays opposing roles in cytotoxicity and interferon-γ production by natural killer and CD8 T cells after influenza A virus infection in mice.
Arimori Y, Nakamura R, Yamada H, Shibata K, Maeda N, Kase T, Yoshikai Y. Arimori Y, et al. J Innate Immun. 2014;6(4):456-66. doi: 10.1159/000356824. Epub 2014 Jan 10. J Innate Immun. 2014. PMID: 24435166 Free PMC article.
Cited by
- Inflammatory monocytes activate memory CD8(+) T and innate NK lymphocytes independent of cognate antigen during microbial pathogen invasion.
Soudja SM, Ruiz AL, Marie JC, Lauvau G. Soudja SM, et al. Immunity. 2012 Sep 21;37(3):549-62. doi: 10.1016/j.immuni.2012.05.029. Epub 2012 Aug 30. Immunity. 2012. PMID: 22940097 Free PMC article. - Hematopoietic Stem Cell Regulation by Type I and II Interferons in the Pathogenesis of Acquired Aplastic Anemia.
Smith JN, Kanwar VS, MacNamara KC. Smith JN, et al. Front Immunol. 2016 Aug 29;7:330. doi: 10.3389/fimmu.2016.00330. eCollection 2016. Front Immunol. 2016. PMID: 27621733 Free PMC article. Review. - Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells.
Behr FM, Chuwonpad A, Stark R, van Gisbergen KPJM. Behr FM, et al. Front Immunol. 2018 Jul 30;9:1770. doi: 10.3389/fimmu.2018.01770. eCollection 2018. Front Immunol. 2018. PMID: 30131803 Free PMC article. Review. - Inflammatory chemokine receptors regulate CD8(+) T cell contraction and memory generation following infection.
Kohlmeier JE, Reiley WW, Perona-Wright G, Freeman ML, Yager EJ, Connor LM, Brincks EL, Cookenham T, Roberts AD, Burkum CE, Sell S, Winslow GM, Blackman MA, Mohrs M, Woodland DL. Kohlmeier JE, et al. J Exp Med. 2011 Aug 1;208(8):1621-34. doi: 10.1084/jem.20102110. Epub 2011 Jul 25. J Exp Med. 2011. PMID: 21788409 Free PMC article. - Environmental cues regulate epigenetic reprogramming of airway-resident memory CD8+ T cells.
Hayward SL, Scharer CD, Cartwright EK, Takamura S, Li ZT, Boss JM, Kohlmeier JE. Hayward SL, et al. Nat Immunol. 2020 Mar;21(3):309-320. doi: 10.1038/s41590-019-0584-x. Epub 2020 Jan 17. Nat Immunol. 2020. PMID: 31953534 Free PMC article.
References
- Borron PJ, Crouch EC, Lewis JF, Wright JR, Possmayer F, Fraher LJ. Recombinant rat surfactant-associated protein D inhibits human T lymphocyte proliferation and IL-2 production. J. Immunol. 1998;161:4599–4603. - PubMed
- Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat. Immunol. 2001;2:1067–1076. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI67967/AI/NIAID NIH HHS/United States
- AI76499/AI/NIAID NIH HHS/United States
- R21 AI083610/AI/NIAID NIH HHS/United States
- R01 AI067967/AI/NIAID NIH HHS/United States
- T32 AI049823/AI/NIAID NIH HHS/United States
- AI83610/AI/NIAID NIH HHS/United States
- T32 AI49823/AI/NIAID NIH HHS/United States
- F32 AI071478/AI/NIAID NIH HHS/United States
- R01 AI076499/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous