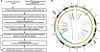SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data - PubMed (original) (raw)
SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data
Bruno Zeitouni et al. Bioinformatics. 2010.
Abstract
Summary: We present SVDetect, a program designed to identify genomic structural variations from paired-end and mate-pair next-generation sequencing data produced by the Illumina GA and ABI SOLiD platforms. Applying both sliding-window and clustering strategies, we use anomalously mapped read pairs provided by current short read aligners to localize genomic rearrangements and classify them according to their type, e.g. large insertions-deletions, inversions, duplications and balanced or unbalanced inter-chromosomal translocations. SVDetect outputs predicted structural variants in various file formats for appropriate graphical visualization.
Availability: Source code and sample data are available at http://svdetect.sourceforge.net/
Figures
Fig. 1.
Overview of SVDetect algorithm and output. (A) The workflow. (B) Graphical visualization of predicted SVs. Genomic locations of inter- and intra-chromosomal links are shown using the Circos software. Starting from outside of the circle, the following features are displayed: chromosome ideograms, scatter plot of the copy-number profile and color-coded spans of chromosomal links.
Similar articles
- SVachra: a tool to identify genomic structural variation in mate pair sequencing data containing inward and outward facing reads.
Hampton OA, English AC, Wang M, Salerno WJ, Liu Y, Muzny DM, Han Y, Wheeler DA, Worley KC, Lupski JR, Gibbs RA. Hampton OA, et al. BMC Genomics. 2017 Oct 3;18(Suppl 6):691. doi: 10.1186/s12864-017-4021-y. BMC Genomics. 2017. PMID: 28984202 Free PMC article. - A streamlined method for detecting structural variants in cancer genomes by short read paired-end sequencing.
Mijušković M, Brown SM, Tang Z, Lindsay CR, Efstathiadis E, Deriano L, Roth DB. Mijušković M, et al. PLoS One. 2012;7(10):e48314. doi: 10.1371/journal.pone.0048314. Epub 2012 Oct 29. PLoS One. 2012. PMID: 23144753 Free PMC article. - RSVSim: an R/Bioconductor package for the simulation of structural variations.
Bartenhagen C, Dugas M. Bartenhagen C, et al. Bioinformatics. 2013 Jul 1;29(13):1679-81. doi: 10.1093/bioinformatics/btt198. Epub 2013 Apr 25. Bioinformatics. 2013. PMID: 23620362 - Geographic distribution and adaptive significance of genomic structural variants: an anthropological genetics perspective.
Eaaswarkhanth M, Pavlidis P, Gokcumen O. Eaaswarkhanth M, et al. Hum Biol. 2014 Fall;86(4):260-75. doi: 10.13110/humanbiology.86.4.0260. Hum Biol. 2014. PMID: 25959693 Review. - Progress in the detection of human genome structural variations.
WU X, XIAO H. WU X, et al. Sci China C Life Sci. 2009 Jun;52(6):560-7. doi: 10.1007/s11427-009-0078-4. Epub 2009 Jun 26. Sci China C Life Sci. 2009. PMID: 19557334 Review.
Cited by
- Bellerophon: a hybrid method for detecting interchromosomal rearrangements at base pair resolution using next-generation sequencing data.
Hayes M, Li J. Hayes M, et al. BMC Bioinformatics. 2013;14 Suppl 5(Suppl 5):S6. doi: 10.1186/1471-2105-14-S5-S6. Epub 2013 Apr 10. BMC Bioinformatics. 2013. PMID: 23734783 Free PMC article. - Partial USH2A deletions contribute to Usher syndrome in Denmark.
Dad S, Rendtorff ND, Kann E, Albrechtsen A, Mehrjouy MM, Bak M, Tommerup N, Tranebjærg L, Rosenberg T, Jensen H, Møller LB. Dad S, et al. Eur J Hum Genet. 2015 Dec;23(12):1646-51. doi: 10.1038/ejhg.2015.54. Epub 2015 Mar 25. Eur J Hum Genet. 2015. PMID: 25804404 Free PMC article. - Breakage-Fusion-Bridge Events Trigger Complex Genome Rearrangements and Amplifications in Developmentally Arrested T Cell Lymphomas.
Bianchi JJ, Murigneux V, Bedora-Faure M, Lescale C, Deriano L. Bianchi JJ, et al. Cell Rep. 2019 Jun 4;27(10):2847-2858.e4. doi: 10.1016/j.celrep.2019.05.014. Cell Rep. 2019. PMID: 31167132 Free PMC article. - Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives.
Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Zhao M, et al. BMC Bioinformatics. 2013;14 Suppl 11(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. Epub 2013 Sep 13. BMC Bioinformatics. 2013. PMID: 24564169 Free PMC article. - DB2: a probabilistic approach for accurate detection of tandem duplication breakpoints using paired-end reads.
Yavaş G, Koyutürk M, Gould MP, McMahon S, LaFramboise T. Yavaş G, et al. BMC Genomics. 2014 Mar 5;15(1):175. doi: 10.1186/1471-2164-15-175. BMC Genomics. 2014. PMID: 24597945 Free PMC article.
References
- Medvedev P, et al. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6(Suppl. 11):S13–S20. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources