Regulation of myeloid leukaemia by the cell-fate determinant Musashi - PubMed (original) (raw)
. 2010 Aug 5;466(7307):765-8.
doi: 10.1038/nature09171. Epub 2010 Jul 18.
Hyog Young Kwon, Bryan Zimdahl, Kendra L Congdon, Jordan Blum, William E Lento, Chen Zhao, Anand Lagoo, Gareth Gerrard, Letizia Foroni, John Goldman, Harriet Goh, Soo-Hyun Kim, Dong-Wook Kim, Charles Chuah, Vivian G Oehler, Jerald P Radich, Craig T Jordan, Tannishtha Reya
Affiliations
- PMID: 20639863
- PMCID: PMC2918284
- DOI: 10.1038/nature09171
Regulation of myeloid leukaemia by the cell-fate determinant Musashi
Takahiro Ito et al. Nature. 2010.
Abstract
Chronic myelogenous leukaemia (CML) can progress from a slow growing chronic phase to an aggressive blast crisis phase, but the molecular basis of this transition remains poorly understood. Here we have used mouse models of CML to show that disease progression is regulated by the Musashi-Numb signalling axis. Specifically, we find that the chronic phase is marked by high levels of Numb expression whereas the blast crisis phase has low levels of Numb expression, and that ectopic expression of Numb promotes differentiation and impairs advanced-phase disease in vivo. As a possible explanation for the decreased levels of Numb in the blast crisis phase, we show that NUP98-HOXA9, an oncogene associated with blast crisis CML, can trigger expression of the RNA-binding protein Musashi2 (Msi2), which in turn represses Numb. Notably, loss of Msi2 restores Numb expression and significantly impairs the development and propagation of blast crisis CML in vitro and in vivo. Finally we show that Msi2 expression is not only highly upregulated during human CML progression but is also an early indicator of poorer prognosis. These data show that the Musashi-Numb pathway can control the differentiation of CML cells, and raise the possibility that targeting this pathway may provide a new strategy for the therapy of aggressive leukaemias.
Figures
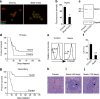
Figure 1. Expression of Numb impairs blast crisis CML development
a, CML cells were immunostained with anti-Numb antibody (red) and DAPI (green pseudocolor) and b, fluorescence intensity quantified *p<0.05. c, CML cells were analyzed by Western blot for Numb expression. d, Cells infected with BCR-ABL, NUP98-HOXA9 and either control vector or Numb were transplanted and survival monitored (control, n=18 and Numb, n=19). e, Representative and f, average frequency of Lin− cells from control or Numb expressing leukemias **p<0.001. g, Lin− cells from primary leukemias were serially transplanted and survival monitored (Vector, n=14 and Numb, n=15, **p<0.001). h, Hematoxylin and Eosin stained spleen sections from control vector or i, Numb expressing leukemias or j, surviving mice. Immature myeloid cells (red arrowheads) and lymphoid follicles (black arrowheads). Magnification: 10×. Error bars in all bar graphs are s.e.m. Data shown is representative of three to four independent experiments.
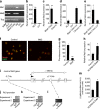
Figure 2. The RNA binding protein Musashi is highly expressed in immature normal and leukemic cells and is regulated by HoxA9
a, Musashi (Msi) expression in whole bone marrow (WBM), KLS cells, chronic and blast crisis CML, olfactory bulb (OB), –reverse transcriptase (-RT in OB) and water. Tbp, TATA binding protein. b–e, Realtime RT-PCR analysis of Msi2 expression in b, KLS cells (n=3), Lin+ cells (n=2) **p<0.001, c, blast crisis phase (n=9), chronic phase (n=6) **p<0.001, d, lin− chronic and blast crisis phase cells relative to normal KLS and lin+ cells (lin+, n=2 and others, n=3), e, lin− (n=5) or lin+ (n=5) blast crisis CML cells *p=0.039. Error bars represent s.e.m. f, Control vector- or Msi2-expressing CML cells were stained with anti-Numb antibody (red) and DAPI (green pseudocolor) and g, fluorescence intensity quantified **p<0.001. h, Msi2 expression in KLS cells transduced with either control vector or NUP98-HOXA9 retrovirus along with BCR-ABL *p=0.017. i–l, HoxA9 binds to the Msi2 promoter. Murine Msi2 gene structure: exons (numbered boxes), transcription start site (TSS; +1) and the direction of transcription (flag), putative HOX binding element 5.7kb upstream of TSS (oval) and +110kb site with no HoxA9 binding sequence (open rectangle). i, ChIP was performed either with IgG control or anti-HoxA9 antibody for j, Flt3, a known HoxA9 target gene, as a positive control and k, Msi2 −5.7kb region or l, Msi2 +110kb region. m, KLS cells from Msi2 genetrap reporter mice were transduced with BCR-ABL with either control vector or NUP98-HOXA9 and β-galactosidase reporter activity quantified (n=2 each, *p=0.011).
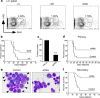
Figure 3. Loss of Musashi impairs the development and propagation of blast crisis CML
a, Frequency of KLS cells in mice of indicated genotypes (+/+, n=4, +/Gt, n=3 and Gt/Gt, n=4). b, Survival curve of mice transplanted with BCR-ABL and NUP98-HOXA9 infected +/+ or Gt/Gt KLS cells (+/+, n=15 and Gt/Gt, n=14, *p=0.0159). c, Colony-forming ability of blast crisis CML cells transduced with control shRNA (shLuc) or Msi2 shRNA (shMsi) **p<0.001. d, Survival curve of mice transplanted with established blast crisis CML cells infected with control shLuc or shMsi (n=13 each, *p=0.0267). e, Wright's stain of leukemic cells from mice transplanted with control shLuc or shMsi infected blast crisis CML. Immature myeloblasts (closed arrowheads), differentiating myelocytes and mature band cells (open arrowheads). Magnification: 100×. f, Survival curve of mice transplanted with Lin− cells from primary shRNA expressing leukemias (Control, n=15 and shMsi, n=16, **p<0.001). Data shown is representative of two to three independent experiments.
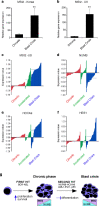
Figure 4. Musashi expression is upregulated during human CML progression
a and b, PCR analysis of MSI2 expression in chronic and blast crisis CML patient samples from a, the Korean Leukemia Bank, Korea (n=9 per cohort, Mann-Whitney U test **p<0.001) and b, the Hammersmith MRD Lab Sample Archive, United Kingdom (n=6 per cohort, Mann-Whitney U test, **p<0.001). c–f, Microarray analysis of expression of c, MSI2 d, NUMB e, HOXA9 (all p<.001) and f, HES1 (p=.68) in bone marrow and peripheral blood samples from 42 chronic (red), 17 accelerated (green) and 31 blast crisis phase (blue) patients in the United States. g, Proposed model for the role of Musashi and Numb in CML progression.
Comment in
- Leukaemia: Comfortably MSI2-NUMB.
Danovi SA. Danovi SA. Nat Rev Cancer. 2010 Sep;10(9):602. doi: 10.1038/nrc2919. Nat Rev Cancer. 2010. PMID: 20803813 No abstract available.
Similar articles
- Aggressive myeloid leukemia formation is directed by the Musashi 2/Numb pathway.
Griner LN, Reuther GW. Griner LN, et al. Cancer Biol Ther. 2010 Nov 15;10(10):979-82. doi: 10.4161/cbt.10.10.14010. Epub 2010 Nov 15. Cancer Biol Ther. 2010. PMID: 21084860 - A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9.
Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, Li S, Van Etten RA, Borrow J, Housman D, Druker B, Gilliland DG. Dash AB, et al. Proc Natl Acad Sci U S A. 2002 May 28;99(11):7622-7. doi: 10.1073/pnas.102583199. Proc Natl Acad Sci U S A. 2002. PMID: 12032333 Free PMC article. - Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia.
Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, Edison AS, Ito T. Hattori A, et al. Nature. 2017 May 25;545(7655):500-504. doi: 10.1038/nature22314. Epub 2017 May 17. Nature. 2017. PMID: 28514443 Free PMC article. - Chronic myelogenous leukemia: mechanisms underlying disease progression.
Shet AS, Jahagirdar BN, Verfaillie CM. Shet AS, et al. Leukemia. 2002 Aug;16(8):1402-11. doi: 10.1038/sj.leu.2402577. Leukemia. 2002. PMID: 12145676 Review. - The molecular biology of chronic myeloid leukaemia.
Melo JV. Melo JV. Leukemia. 1996 May;10(5):751-6. Leukemia. 1996. PMID: 8656667 Review.
Cited by
- Functional Integration of mRNA Translational Control Programs.
MacNicol MC, Cragle CE, Arumugam K, Fosso B, Pesole G, MacNicol AM. MacNicol MC, et al. Biomolecules. 2015 Jul 21;5(3):1580-99. doi: 10.3390/biom5031580. Biomolecules. 2015. PMID: 26197342 Free PMC article. Review. - Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration.
Bennett CG, Riemondy K, Chapnick DA, Bunker E, Liu X, Kuersten S, Yi R. Bennett CG, et al. Nucleic Acids Res. 2016 May 5;44(8):3788-800. doi: 10.1093/nar/gkw207. Epub 2016 Mar 31. Nucleic Acids Res. 2016. PMID: 27034466 Free PMC article. - Declaration of Bcr-Abl1 independence.
Zhao H, Deininger MW. Zhao H, et al. Leukemia. 2020 Nov;34(11):2827-2836. doi: 10.1038/s41375-020-01037-9. Epub 2020 Sep 10. Leukemia. 2020. PMID: 32908250 Review. No abstract available. - Sox2: regulation of expression and contribution to brain tumors.
Mansouri S, Nejad R, Karabork M, Ekinci C, Solaroglu I, Aldape KD, Zadeh G. Mansouri S, et al. CNS Oncol. 2016 Jul;5(3):159-73. doi: 10.2217/cns-2016-0001. Epub 2016 May 27. CNS Oncol. 2016. PMID: 27230973 Free PMC article. Review. - Musashi2 is required for the self-renewal and pluripotency of embryonic stem cells.
Wuebben EL, Mallanna SK, Cox JL, Rizzino A. Wuebben EL, et al. PLoS One. 2012;7(4):e34827. doi: 10.1371/journal.pone.0034827. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496868 Free PMC article.
References
- Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. - PubMed
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. - PubMed
- Pear WS, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. - PubMed
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. - PubMed
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK072234-04/DK/NIDDK NIH HHS/United States
- R01 CA140371/CA/NCI NIH HHS/United States
- T32 GM007184/GM/NIGMS NIH HHS/United States
- DP1 OD006430/OD/NIH HHS/United States
- R01 DK063031-07/DK/NIDDK NIH HHS/United States
- R01 HL097767/HL/NHLBI NIH HHS/United States
- R01 HL097767-01/HL/NHLBI NIH HHS/United States
- R01 DK063031/DK/NIDDK NIH HHS/United States
- R01 DK063031-01S1/DK/NIDDK NIH HHS/United States
- T32 GM007184-33/GM/NIGMS NIH HHS/United States
- R01 CA122206/CA/NCI NIH HHS/United States
- DK072234/DK/NIDDK NIH HHS/United States
- CA140371/CA/NCI NIH HHS/United States
- CA18029/CA/NCI NIH HHS/United States
- R01 DK063031-03/DK/NIDDK NIH HHS/United States
- R01 DK072234-01A1/DK/NIDDK NIH HHS/United States
- P01 CA018029/CA/NCI NIH HHS/United States
- HL097767/HL/NHLBI NIH HHS/United States
- U19 AI067798-040006/AI/NIAID NIH HHS/United States
- R01 DK063031-07S1/DK/NIDDK NIH HHS/United States
- DP1 OD006430-01/OD/NIH HHS/United States
- R01 DK072234/DK/NIDDK NIH HHS/United States
- R01 DK063031-08/DK/NIDDK NIH HHS/United States
- R01 DK063031-02/DK/NIDDK NIH HHS/United States
- U19 AI067798-010006/AI/NIAID NIH HHS/United States
- AI067798/AI/NIAID NIH HHS/United States
- DP1OD006430/OD/NIH HHS/United States
- DP1 OD006430-02/OD/NIH HHS/United States
- U19 AI067798-020006/AI/NIAID NIH HHS/United States
- R01 DK063031-05/DK/NIDDK NIH HHS/United States
- DP1 CA174422/CA/NCI NIH HHS/United States
- U19 AI067798-050006/AI/NIAID NIH HHS/United States
- R01 DK063031-01/DK/NIDDK NIH HHS/United States
- DK63031/DK/NIDDK NIH HHS/United States
- CA122206/CA/NCI NIH HHS/United States
- R01 DK063031-06/DK/NIDDK NIH HHS/United States
- R01 HL097767-02/HL/NHLBI NIH HHS/United States
- U19 AI067798/AI/NIAID NIH HHS/United States
- R01 DK072234-02/DK/NIDDK NIH HHS/United States
- R01 DK063031-04/DK/NIDDK NIH HHS/United States
- U19 AI067798-030006/AI/NIAID NIH HHS/United States
- R01 DK072234-03/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous