Epigenetic memory in induced pluripotent stem cells - PubMed (original) (raw)
. 2010 Sep 16;467(7313):285-90.
doi: 10.1038/nature09342.
A Doi, B Wen, K Ng, R Zhao, P Cahan, J Kim, M J Aryee, H Ji, L I R Ehrlich, A Yabuuchi, A Takeuchi, K C Cunniff, H Hongguang, S McKinney-Freeman, O Naveiras, T J Yoon, R A Irizarry, N Jung, J Seita, J Hanna, P Murakami, R Jaenisch, R Weissleder, S H Orkin, I L Weissman, A P Feinberg, G Q Daley
Affiliations
- PMID: 20644535
- PMCID: PMC3150836
- DOI: 10.1038/nature09342
Epigenetic memory in induced pluripotent stem cells
K Kim et al. Nature. 2010.
Abstract
Somatic cell nuclear transfer and transcription-factor-based reprogramming revert adult cells to an embryonic state, and yield pluripotent stem cells that can generate all tissues. Through different mechanisms and kinetics, these two reprogramming methods reset genomic methylation, an epigenetic modification of DNA that influences gene expression, leading us to hypothesize that the resulting pluripotent stem cells might have different properties. Here we observe that low-passage induced pluripotent stem cells (iPSCs) derived by factor-based reprogramming of adult murine tissues harbour residual DNA methylation signatures characteristic of their somatic tissue of origin, which favours their differentiation along lineages related to the donor cell, while restricting alternative cell fates. Such an 'epigenetic memory' of the donor tissue could be reset by differentiation and serial reprogramming, or by treatment of iPSCs with chromatin-modifying drugs. In contrast, the differentiation and methylation of nuclear-transfer-derived pluripotent stem cells were more similar to classical embryonic stem cells than were iPSCs. Our data indicate that nuclear transfer is more effective at establishing the ground state of pluripotency than factor-based reprogramming, which can leave an epigenetic memory of the tissue of origin that may influence efforts at directed differentiation for applications in disease modelling or treatment.
Figures
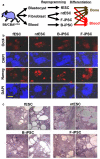
Figure 1. Pluripotent stem cells and their characterization
a, Experimental schema. fESC, ntESC, F-iPSC, and B-iPSC were derived from B6/CBA F1 mice by reprogramming and/or cell culture, characterized for pluripotency by criteria applied to human cells, followed by differentiation analysis for osteogenic or hematopoietic lineages. b, Expression of pluripotency markers NANOG and OCT4 by immunohistochemistry. 4,6-Diamidino-2-phenylindole (DAPI) staining for total cell content. Feeder fibroblasts serve as internal negative controls. c, Teratoma analysis: tumor histology from indicated cell lines shows highly cystic structures consisting of differentiated elements of all three embryonic germ layers. Scale bar, 200μm.
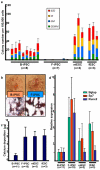
Figure 2. Differentiation of cell lines
a, Hematopoietic colony number per 100,000 EB cells differentiated from indicated cell lines. b, Alizarin Red staining of osteogenic cultures of B-iPSC (left) and F-iPSC (right). Top: 3 cm culture dish; Bottom: stained colonies; scale bar, 500 μm. c, Quantification of elemental calcium by inductively coupled plasma – atomic emission spectroscopy in 5×105 cells after osteogenic differentiation of indicated cell lines. d, Q-PCR of osteogenic genes, Bglap, Sp7, and Runx2 in indicated cell lines after osteogenic differentiation. Gene expression was normalized to Actin. n=number of independent clones tested. Error bars=s.d.
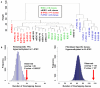
Figure 3. Analysis of methylation in stem cell lines
a, Cluster dendrogram using probes from DMRs that distinguish B-iPSC and F-iPSC. Cell clones are described in Supplemental Table 5. b, Enrichment of DMRs for hematopoiesis and fibroblast-related transcription factors in B-iPSC and F-iPSC, relative to chance (blue histogram; 100,000 random permutations). Left panel: 20 of 74 hematopoiesis-related transcription factors overlap DMRs hypermethylated in F-IPSC (p=0.0034); Right panel: 115 of 764 fibroblast-specific genes overlap DMRs hypermethylated in B-iPSC (p=10−5).
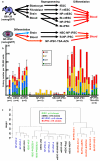
Figure 4. Stringently-defined pluripotent stem cells and their characterization
a, Experimental schema. Four horizontal lines indicate integrated proviruses carrying dox-inducible reprogramming factors in some experiments. Characteristics of individual clones in all subsequent panels can be found in Supplemental Table 5. b, Hematopoietic colony number per 100,000 EB cells differentiated from indicated cell lines. n=number of independent clones tested. Error bars=s.d., added for clones repeated three or more times. c, Cluster dendrogram using probes from DMRs that distinguish Bl-iPSC and NP-iPSC.
Comment in
- Stem cells: Troublesome memories.
Zwaka TP. Zwaka TP. Nature. 2010 Sep 16;467(7313):280-1. doi: 10.1038/467280a. Nature. 2010. PMID: 20844526 No abstract available.
Similar articles
- Stem cells: Troublesome memories.
Zwaka TP. Zwaka TP. Nature. 2010 Sep 16;467(7313):280-1. doi: 10.1038/467280a. Nature. 2010. PMID: 20844526 No abstract available. - Integrated analysis of hematopoietic differentiation outcomes and molecular characterization reveals unbiased differentiation capacity and minor transcriptional memory in HPC/HSC-iPSCs.
Gao S, Hou X, Jiang Y, Xu Z, Cai T, Chen J, Chang G. Gao S, et al. Stem Cell Res Ther. 2017 Jan 23;8(1):13. doi: 10.1186/s13287-016-0466-1. Stem Cell Res Ther. 2017. PMID: 28114969 Free PMC article. - High throughput sequencing identifies an imprinted gene, Grb10, associated with the pluripotency state in nuclear transfer embryonic stem cells.
Li H, Gao S, Huang H, Liu W, Huang H, Liu X, Gao Y, Le R, Kou X, Zhao Y, Kou Z, Li J, Wang H, Zhang Y, Wang H, Cai T, Sun Q, Gao S, Han Z. Li H, et al. Oncotarget. 2017 Jul 18;8(29):47344-47355. doi: 10.18632/oncotarget.17185. Oncotarget. 2017. PMID: 28476045 Free PMC article. - Epigenetics of induced pluripotency, the seven-headed dragon.
Djuric U, Ellis J. Djuric U, et al. Stem Cell Res Ther. 2010 Mar 15;1(1):3. doi: 10.1186/scrt3. Stem Cell Res Ther. 2010. PMID: 20504284 Free PMC article. Review. - Concise review: Induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart?
Bilic J, Izpisua Belmonte JC. Bilic J, et al. Stem Cells. 2012 Jan;30(1):33-41. doi: 10.1002/stem.700. Stem Cells. 2012. PMID: 22213481 Review.
Cited by
- Cardiovascular repair with bone marrow-derived cells.
Kim WS, Lee S, Yoon YS. Kim WS, et al. Blood Res. 2013 Jun;48(2):76-86. doi: 10.5045/br.2013.48.2.76. Epub 2013 Jun 25. Blood Res. 2013. PMID: 23826576 Free PMC article. - Cardiac progenitors derived from reprogrammed mesenchymal stem cells contribute to angiomyogenic repair of the infarcted heart.
Buccini S, Haider KH, Ahmed RP, Jiang S, Ashraf M. Buccini S, et al. Basic Res Cardiol. 2012 Nov;107(6):301. doi: 10.1007/s00395-012-0301-5. Epub 2012 Oct 18. Basic Res Cardiol. 2012. PMID: 23076626 Free PMC article. - HLA engineering of human pluripotent stem cells.
Riolobos L, Hirata RK, Turtle CJ, Wang PR, Gornalusse GG, Zavajlevski M, Riddell SR, Russell DW. Riolobos L, et al. Mol Ther. 2013 Jun;21(6):1232-41. doi: 10.1038/mt.2013.59. Epub 2013 Apr 30. Mol Ther. 2013. PMID: 23629003 Free PMC article. - The effect of age on stem cell function and utility for therapy.
Narbonne P. Narbonne P. Cell Med. 2018 Jun 8;10:2155179018773756. doi: 10.1177/2155179018773756. eCollection 2018. Cell Med. 2018. PMID: 32634187 Free PMC article. Review. - Post-natal cardiomyocytes can generate iPS cells with an enhanced capacity toward cardiomyogenic re-differentation.
Rizzi R, Di Pasquale E, Portararo P, Papait R, Cattaneo P, Latronico MV, Altomare C, Sala L, Zaza A, Hirsch E, Naldini L, Condorelli G, Bearzi C. Rizzi R, et al. Cell Death Differ. 2012 Jul;19(7):1162-74. doi: 10.1038/cdd.2011.205. Epub 2012 Jan 20. Cell Death Differ. 2012. PMID: 22261617 Free PMC article.
References
Method Reference
- Kirov G, et al. Variation in the protocadherin gamma A gene cluster. Genomics. 2003;82:433–440. - PubMed
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. Epub 2006 Aug 2010. - PubMed
- Zhao XY, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. - PubMed
- Daley GQ, et al. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–201. author reply 202. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL099999/HL/NHLBI NIH HHS/United States
- R01AI047458/AI/NIAID NIH HHS/United States
- R01-DK70055/DK/NIDDK NIH HHS/United States
- R01 DK059279/DK/NIDDK NIH HHS/United States
- R37 CA054358/CA/NCI NIH HHS/United States
- P50 HG003233/HG/NHGRI NIH HHS/United States
- RC2-HL102815/HL/NHLBI NIH HHS/United States
- R01 DK070055/DK/NIDDK NIH HHS/United States
- CA86065/CA/NCI NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- K99 HL093212/HL/NHLBI NIH HHS/United States
- R37CA054358/CA/NCI NIH HHS/United States
- R01AI047457/AI/NIAID NIH HHS/United States
- P50HG003233/HG/NHGRI NIH HHS/United States
- R01 AI047457/AI/NIAID NIH HHS/United States
- R01-DK59279/DK/NIDDK NIH HHS/United States
- R01 CA086065/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 HL099999/HL/NHLBI NIH HHS/United States
- K99HL093212-01/HL/NHLBI NIH HHS/United States
- R01 GM083084/GM/NIGMS NIH HHS/United States
- R01 AI047458/AI/NIAID NIH HHS/United States
- DP1 OD000256/OD/NIH HHS/United States
- RC2 HL102815/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases