Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells - PubMed (original) (raw)
doi: 10.1038/nbt.1667. Epub 2010 Jul 19.
Susanna Liu, Maria Eugenia Figueroa, Warakorn Kulalert, Sarah Eminli, Kah Yong Tan, Effie Apostolou, Matthias Stadtfeld, Yushan Li, Toshi Shioda, Sridaran Natesan, Amy J Wagers, Ari Melnick, Todd Evans, Konrad Hochedlinger
Affiliations
- PMID: 20644536
- PMCID: PMC3148605
- DOI: 10.1038/nbt.1667
Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells
Jose M Polo et al. Nat Biotechnol. 2010 Aug.
Abstract
Induced pluripotent stem cells (iPSCs) have been derived from various somatic cell populations through ectopic expression of defined factors. It remains unclear whether iPSCs generated from different cell types are molecularly and functionally similar. Here we show that iPSCs obtained from mouse fibroblasts, hematopoietic and myogenic cells exhibit distinct transcriptional and epigenetic patterns. Moreover, we demonstrate that cellular origin influences the in vitro differentiation potentials of iPSCs into embryoid bodies and different hematopoietic cell types. Notably, continuous passaging of iPSCs largely attenuates these differences. Our results suggest that early-passage iPSCs retain a transient epigenetic memory of their somatic cells of origin, which manifests as differential gene expression and altered differentiation capacity. These observations may influence ongoing attempts to use iPSCs for disease modeling and could also be exploited in potential therapeutic applications to enhance differentiation into desired cell lineages.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/.
Figures
Figure 1
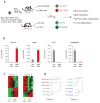
iPSCs derived from different cell types are transcriptionally distinguishable. (a) Flow chart explaining the derivation and analysis of genetically matched iPSCs from different cell types. Secondary iPSCs were first injected into blastocysts to generate chimeric mice, from which the indicated somatic cell types were isolated. Exposure of these cells to doxycycline (dox) then gave rise to iPSCs. ChIP, chromatin immunoprecipitation. (b) Quantification of the expression levels of Cxcr4, Itgb1, Gr-1 and Lysozyme by quantitative PCR in SMP-iPSCs, in red, and Gra-iPSCs, in gray. The values were normalized to GAPDH expression; the error bars depict the s.e.m. (n = 3). (c) Heat map showing top 104 probes with highest variance in their expression levels. Left panel, SMP-iPSCs and Gra-iPSCs derived from chimera no. 1. Right panel, TTF-iPSCs and B-iPSCs derived from chimera no. 2. (d) Hierarchical, unsupervised clustering of iPSC expression profiles using the correlation distance and the Ward method. SMP-iPSCs and Gra-iPSCs were derived from chimera no. 1 (left panel), TTF-iPSCs and B-iPSCs originate from chimera no. 2 (right panel). Chi no. 1, chimera no. 1; chi no. 2, chimera no. 2.
Figure 2
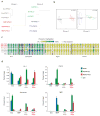
iPSCs derived from different cell types exhibit distinguishable epigenetic signatures. (a) Hierarchical unsupervised clustering analysis of HELP genome-wide methylation data from indicated iPSC lines. (b) Correspondence analysis of SMP-iPSCs and Gra-iPSCs (left panel) from chimera no. 1, TTF-iPSCs and B-iPSCs (right panel) from chimera no. 2. (c) Graphic representation of DNA methylation quantification of specific CpGs (circles) in the promoter regions of the indicated candidate genes using EpiTYPER DNA methylation analyses. Yellow indicates 0% methylation and blue 100% methylation. (d) Chromatin immunoprecipitation (ChIP) for H3 pan-acetylated (H3Ac, in blue), H3K4 trimethylated (H3K4me3, in green), H3K27 trimethylated (H3K27me3, in red) and isotype control (IgG, in light blue) of granulocytes (Gra), SMPs, Gra-iPSCs and SMP-iPSCs. Chi no. 1, chimera no. 1; chi no. 2, chimera no. 2. The error bars depict the s.e.m. (n = 3).
Figure 3
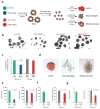
iPSCs derived from different cell types have distinctive in vitro differentiation potentials. (a) experimental outline. iPSCs were first differentiated into embryoid bodies. At day 6, embryoid bodies were dissociated and plated in conditions to favor differentiation into erythrocyte progenitors (eryP) and macrophage and mixed hematopoietic colonies. (b) Phase contrast images showing embryoid bodies derived from B-iPSCs, TTF-iPSCs, Gra-iPSCs and SMP-iPSCs at same magnification. (c) Quantification of embryoid body sizes derived from B-iPSCs, TTF-iPSCs, Gra-iPSCs and SMP-iPSCs; the diameter of the embryoid bodies was measured using arbitrary units (AU). The error bars depict the s.e.m. (n = 30) (d) Representative images of erythrocyte progenitors (eryPs), macrophage colonies and mixed hematopoietic colonies. (e–g) Quantification of in vitro differentiation potentials of the different iPSCs into EryPs (e), macrophage colonies (f) and mixed hematopoietic colonies (g). Chi no. 1, chimera no. 1; chi no. 2, chimera no. 2. The error bars depict the s.e.m. (n = 12).
Figure 4
Continuous passaging of iPSCs abrogates transcriptional, epigenetic and functional differences. (a) Hierarchical unsupervised clustering of expression profiles from B-iPSCs, T-iPSCs, TTF-iPSCs and Gra-iPSCs from chimera no. 2. Left panel shows clustering analysis of all iPSC samples at passage p4, the middle panel at p10 and the right panel at p16. (b) Number of differentially expressed probes between pairs of iPSC samples used in a; iPSCs at p4 are shown in blue bars, iPSCs at p10 are shown in orange bars and iPSCs at p16 are shown in red bars. The number of differently expressed probes between iPSCs was calculated using a pairwise analysis (twofold), with _t_-test P = 0.05, with Bejamini and Hochberg correction (n = 3). (c) Venn diagram and GO analysis showing overlap of genes that change from p4 to p16 in Gra-IPSCs, TTF-iPSCs and B-iPSCs. Red line marks functional GO cluster of genes shared between all three iPSC groups. Black line marks functional GO cluster of genes shared by at least two of the iPSC groups. Functional ontology cluster analysis was performed using the DAVIS algorithm. (d) Hierarchical unsupervised clustering using HELP genome-wide methylation profiles of B-iPSCs and TTF-iPSCs at p16. (e–g) Quantification of in vitro differentiation potentials of B-iPSCs and TTF-iPSCs at p16 into EryPs (e), macrophage colonies (f) and mixed hematopoietic colonies (g). The error bars depict the s.e.m. (n = 9).
Figure 5
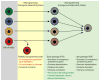
Model summarizing the presented data. iPSCs derived from different somatic cell types retain a transient epigenetic and transcriptional memory of their cell type of origin at early passage, despite acquiring pluripotent gene expression, transgene-independent growth and the ability to contribute to tissues in chimeras. Continuous passaging resolves these differences, giving rise to iPSCs that are molecularly and functionally indistinguishable. Note the difference between early passage iPSCs and partially reprogrammed cells, which require continuous viral transgene expression and fail to activate endogenous pluripotency genes or support the development of viable mice.
Comment in
- Stem cells: holding on to the memories.
Baumann K. Baumann K. Nat Rev Mol Cell Biol. 2010 Sep;11(9):601. doi: 10.1038/nrm2962. Epub 2010 Aug 18. Nat Rev Mol Cell Biol. 2010. PMID: 20717148 No abstract available. - Stem cells: Holding onto the memories.
Baumann K. Baumann K. Nat Rev Genet. 2010 Sep;11(9):593. doi: 10.1038/nrg2849. Nat Rev Genet. 2010. PMID: 20717151 No abstract available.
Similar articles
- Efficient hematopoietic redifferentiation of induced pluripotent stem cells derived from primitive murine bone marrow cells.
Pfaff N, Lachmann N, Kohlscheen S, Sgodda M, Araúzo-Bravo MJ, Greber B, Kues W, Glage S, Baum C, Niemann H, Schambach A, Cantz T, Moritz T. Pfaff N, et al. Stem Cells Dev. 2012 Mar 20;21(5):689-701. doi: 10.1089/scd.2011.0010. Epub 2011 Aug 24. Stem Cells Dev. 2012. PMID: 21732815 - MicroRNAs contribute to induced pluripotent stem cell somatic donor memory.
Vitaloni M, Pulecio J, Bilic J, Kuebler B, Laricchia-Robbio L, Izpisua Belmonte JC. Vitaloni M, et al. J Biol Chem. 2014 Jan 24;289(4):2084-98. doi: 10.1074/jbc.M113.538702. Epub 2013 Dec 5. J Biol Chem. 2014. PMID: 24311783 Free PMC article. - Variation in mesodermal and hematopoietic potential of adult skin-derived induced pluripotent stem cell lines in mice.
Inoue T, Kulkeaw K, Okayama S, Tani K, Sugiyama D. Inoue T, et al. Stem Cell Rev Rep. 2011 Nov;7(4):958-68. doi: 10.1007/s12015-011-9249-3. Stem Cell Rev Rep. 2011. PMID: 21424235 - Hematopoietic cell differentiation from embryonic and induced pluripotent stem cells.
Lim WF, Inoue-Yokoo T, Tan KS, Lai MI, Sugiyama D. Lim WF, et al. Stem Cell Res Ther. 2013 Jun 18;4(3):71. doi: 10.1186/scrt222. Stem Cell Res Ther. 2013. PMID: 23796405 Free PMC article. Review. - Generating autologous hematopoietic cells from human-induced pluripotent stem cells through ectopic expression of transcription factors.
Hwang Y, Broxmeyer HE, Lee MR. Hwang Y, et al. Curr Opin Hematol. 2017 Jul;24(4):283-288. doi: 10.1097/MOH.0000000000000343. Curr Opin Hematol. 2017. PMID: 28383341 Review.
Cited by
- Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells.
Thatava T, Kudva YC, Edukulla R, Squillace K, De Lamo JG, Khan YK, Sakuma T, Ohmine S, Terzic A, Ikeda Y. Thatava T, et al. Mol Ther. 2013 Jan;21(1):228-39. doi: 10.1038/mt.2012.245. Epub 2012 Nov 27. Mol Ther. 2013. PMID: 23183535 Free PMC article. - Technological overview of iPS induction from human adult somatic cells.
Bayart E, Cohen-Haguenauer O. Bayart E, et al. Curr Gene Ther. 2013 Apr;13(2):73-92. doi: 10.2174/1566523211313020002. Curr Gene Ther. 2013. PMID: 23320476 Free PMC article. Review. - Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency.
Studer L, Vera E, Cornacchia D. Studer L, et al. Cell Stem Cell. 2015 Jun 4;16(6):591-600. doi: 10.1016/j.stem.2015.05.004. Cell Stem Cell. 2015. PMID: 26046759 Free PMC article. Review. - Tissue-Specific Stem Cells Obtained by Reprogramming of Non-Obese Diabetic (NOD) Mouse-Derived Pancreatic Cells Confer Insulin Production in Response to Glucose.
Saitoh I, Sato M, Soda M, Inada E, Iwase Y, Murakami T, Ohshima H, Hayasaki H, Noguchi H. Saitoh I, et al. PLoS One. 2016 Sep 23;11(9):e0163580. doi: 10.1371/journal.pone.0163580. eCollection 2016. PLoS One. 2016. PMID: 27662374 Free PMC article. - Reestablishment of the inactive X chromosome to the ground state through cell fusion-induced reprogramming.
Choi HW, Kim JS, Jang HJ, Choi S, Kim JH, Schöler HR, Do JT. Choi HW, et al. Cell Mol Life Sci. 2012 Dec;69(23):4067-77. doi: 10.1007/s00018-012-1139-6. Epub 2012 Nov 8. Cell Mol Life Sci. 2012. PMID: 26250157 Free PMC article.
References
- Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. - PubMed
- Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. - PubMed
Publication types
MeSH terms
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- R01 HL056182/HL/NHLBI NIH HHS/United States
- P30 DK036836/DK/NIDDK NIH HHS/United States
- DP2 OD004345/OD/NIH HHS/United States
- HL056182/HL/NHLBI NIH HHS/United States
- R37 HL056182/HL/NHLBI NIH HHS/United States
- R01 HD058013/HD/NICHD NIH HHS/United States
- P30DK036836/DK/NIDDK NIH HHS/United States
- DP2 OD004345-01/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases