Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer's disease progression - PubMed (original) (raw)
Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer's disease progression
Stephen D Ginsberg et al. Biol Psychiatry. 2010.
Abstract
Background: Endocytic dysfunction and neurotrophin signaling deficits may underlie the selective vulnerability of hippocampal neurons during the progression of Alzheimer's disease (AD), although there is little direct in vivo and biochemical evidence to support this hypothesis.
Methods: Microarray analysis of hippocampal CA1 pyramidal neurons acquired via laser capture microdissection was performed using postmortem brain tissue. Validation was achieved using real-time quantitative polymerase chain reaction and immunoblot analysis. Mechanistic studies were performed using human fibroblasts subjected to overexpression with viral vectors or knockdown via small interference RNA.
Results: Expression levels of genes regulating early endosomes (rab5) and late endosomes (rab7) are selectively upregulated in homogeneous populations of CA1 neurons from individuals with mild cognitive impairment and AD. The levels of these genes are selectively increased as antemortem measures of cognition decline during AD progression. Hippocampal quantitative polymerase chain reaction and immunoblot analyses confirmed increased levels of these transcripts and their respective protein products. Elevation of select rab GTPases regulating endocytosis paralleled the downregulation of genes encoding the neurotrophin receptors TrkB and TrkC. Overexpression of rab5 in cells suppressed TrkB expression, whereas knockdown of TrkB expression did not alter rab5 levels, suggesting that TrkB downregulation is a consequence of endosomal dysfunction associated with elevated rab5 levels in early AD.
Conclusions: These data support the hypothesis that neuronal endosomal dysfunction is associated with preclinical AD. Increased endocytic pathway activity, driven by elevated rab GTPase expression, may result in long-term deficits in hippocampal neurotrophic signaling and represent a key pathogenic mechanism underlying AD progression.
Copyright © 2010 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
TC RNA amplification procedure and LCM of CA1 hippocampal neurons. A. Schematic overview of the TC RNA amplification procedure. mRNA extracted from LCM captured cells (green line) and the TC primer serve as templates for the first strand synthesis with poly d(T) acting as a primer. First strand cDNA consists of three portions, the 5′ end comprised of the poly d(T), the mRNA complementary portion in the middle (purple line), and the 3′ end is comprised of the TC primer complementary to the cDNA (denoted as TC′). The TC′ portion hybridizes with the TC primer present in the reaction and forms a double stranded region which provides a functional RNA synthesis promoter for in vitro transcription and robust RNA amplification. B. Neurofilament-immunoreactive CA1 neurons in the hippocampal pyramidal cell layer from a subject with AD prior to LCM. Tissue sections were dehydrated but not coverslipped to enable proper execution of the LCM process. Scale bar: 50 mm. C. Slightly larger image in B following LCM of an individual CA1 neuron. Scale bar: 50 mm.
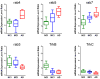
Figure 2
Differential regulation of select rab GTPases and neurotrophin receptors during the progression of AD. Boxplots (NCI, green; MCI, blue; AD, red) indicating relative expression levels of select rab GTPases and neurotrophin receptors. Significant up regulation of rab4, rab5, and rab7 is depicted along with significant down regulation of rab3, TrkB, and TrkC.
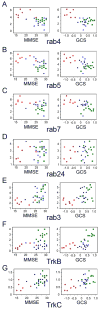
Figure 3
Association between select rab GTPase and neurotrophin receptor gene expression levels within CA1 neurons and antemortem cognitive measures in the same subjects. Scatterplots illustrate the relationship of gene expression levels and MMSE (left panel) and GCS scores (right panel) from individuals classified as NCI (green squares), MCI (blue triangles), and AD (red circles). A. A highly significant negative association exists with rab4 and MMSE (p < 0.0001) and GCS performance (p < 0.0002). B. A highly significant negative association exists with rab5 and MMSE (p < 0.001) and GCS performance (p < 0.001). C. A significant negative association exists with rab7 and MMSE (p < 0.01) and GCS performance (p < 0.001). D. rab24 expression levels are inversely correlated with MMSE (p < 0.0004) and GCS (p < 0.0002) scores. E. A significant association between rab3 gene expression down regulation and poor MMSE (p < 0.002) and GCS (p < 0.005) performance was observed. F. Down regulation of TrkB expression correlated with MMSE (p < 0.02) and GCS (p < 0.0001) performance. G. Down regulation of TrkC expression correlated with MMSE (p < 0.003) and GCS (p < 0.0004) performance.
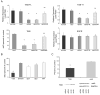
Figure 4
Histograms showing expression of TrkB and EGFR levels in fibroblasts infected with rab5 viral vector constructs and expression of select rab GTPases following TrkB siRNA in cultured human fibroblasts. A. Upper left panel; down regulation of TrkB-FL is observed in fibroblasts infected with rab5wt (single asterisk denotes p < 0.005) and the constitutively active construct rab5Q79L (asterisk) compared to mock transfected (sucrose) control (LacZ) transfected cells. TrkB-FL down regulation was also found in the rab5wt and rabQ79L constructs compared to the dominant negative construct rab5S34N (double asterisk denotes p < 0.01). Upper right panel; down regulation of TrkB-T1 is seen in fibroblasts infected with rab5wt (single asterisk denotes p < 0.007) and the constitutively active construct rab5Q79L (asterisk) compared to mock transfected (sucrose) control (LacZ) transfected cells. Similar to TrkB-FL, TrkB-T1 down regulation was observed in the rab5wt and rabQ79L constructs compared to rab5S34N (double asterisk denotes p < 0.02). Lower left panel; down regulation of TrkB is observed via qPCR in fibroblasts infected with rab5wt and rab5Q79L (single asterisk denotes p < 0.001) compared to control (LacZ) infected cells and to the dominant negative rab5S34N construct (double asterisk denotes p < 0.04), suggesting that rab5 overexpression regulates TrkB at the transcriptional level. Lower right panel; in contrast to differential TrkB expression levels, no significant differences in related EGFR expression were observed with any of the fibroblasts infected with rab5 constructs or controls. TrkB and EGFR levels were normalized to TUBB ± SD for quantitative immunoblot analysis. B. qPCR analysis following TrkB siRNA (Silencer 753) indicated significant knockdown of TrkB expression {34.4% ± 0.9 (SEM); p < 0.001; asterisk} compared to vector treated cells. In contrast, no significant regulation of Gapdh (118.8% ± 7.8), rab4 (86.4% ± 7.8), rab5 (98.5% ± 11.5), or rab7 (114.9% ± 9.3) levels was observed following TrkB siRNA. C. Upper panel; immunoblot analysis demonstrated significant knockdown of TrkB expression (56.4% ± 4.1 (SEM); p < 0.005; asterisk), but not rab5 expression (95.0% ± 3.0), following TrkB siRNA. Lower panel; representative immunoblots demonstrating knockdown of TrkB expression with no significant alterations in rab5 or GAPDH expression.
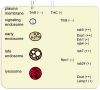
Figure 5
Schematic diagram illustrating gene expression level changes within components of the endosomal-lysosomal system and neurotrophins at the cell surface and within signaling endosomes of vulnerable CA1 pyramidal neurons in MCI and AD. Genes demonstrating up regulation are denoted with a plus sign (one plus sign for ‘late’ changes observed only in AD and two plus signs indicating ‘early’ up regulation seen in MCI and AD). Genes demonstrating down regulation are denoted with a minus sign (one minus sign for ‘late’ changes observed only in AD and two minus signs indicating ‘early’ up regulation seen in MCI and AD).
Similar articles
- Selective decline of neurotrophin and neurotrophin receptor genes within CA1 pyramidal neurons and hippocampus proper: Correlation with cognitive performance and neuropathology in mild cognitive impairment and Alzheimer's disease.
Ginsberg SD, Malek-Ahmadi MH, Alldred MJ, Che S, Elarova I, Chen Y, Jeanneteau F, Kranz TM, Chao MV, Counts SE, Mufson EJ. Ginsberg SD, et al. Hippocampus. 2019 May;29(5):422-439. doi: 10.1002/hipo.22802. Epub 2017 Sep 27. Hippocampus. 2019. PMID: 28888073 Free PMC article. - Upregulation of select rab GTPases in cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer's disease.
Ginsberg SD, Mufson EJ, Alldred MJ, Counts SE, Wuu J, Nixon RA, Che S. Ginsberg SD, et al. J Chem Neuroanat. 2011 Oct;42(2):102-10. doi: 10.1016/j.jchemneu.2011.05.012. Epub 2011 Jun 12. J Chem Neuroanat. 2011. PMID: 21669283 Free PMC article. - Gene expression levels assessed by CA1 pyramidal neuron and regional hippocampal dissections in Alzheimer's disease.
Ginsberg SD, Alldred MJ, Che S. Ginsberg SD, et al. Neurobiol Dis. 2012 Jan;45(1):99-107. doi: 10.1016/j.nbd.2011.07.013. Epub 2011 Jul 28. Neurobiol Dis. 2012. PMID: 21821124 Free PMC article. - Dysregulation of Rab5-mediated endocytic pathways in Alzheimer's disease.
Xu W, Fang F, Ding J, Wu C. Xu W, et al. Traffic. 2018 Apr;19(4):253-262. doi: 10.1111/tra.12547. Epub 2018 Feb 5. Traffic. 2018. PMID: 29314494 Free PMC article. Review. - The Rab11-regulated endocytic pathway and BDNF/TrkB signaling: Roles in plasticity changes and neurodegenerative diseases.
Moya-Alvarado G, Guerra MV, Tiburcio R, Bravo E, Bronfman FC. Moya-Alvarado G, et al. Neurobiol Dis. 2022 Sep;171:105796. doi: 10.1016/j.nbd.2022.105796. Epub 2022 Jun 18. Neurobiol Dis. 2022. PMID: 35728773 Review.
Cited by
- Hippocampal proNGF signaling pathways and β-amyloid levels in mild cognitive impairment and Alzheimer disease.
Mufson EJ, He B, Nadeem M, Perez SE, Counts SE, Leurgans S, Fritz J, Lah J, Ginsberg SD, Wuu J, Scheff SW. Mufson EJ, et al. J Neuropathol Exp Neurol. 2012 Nov;71(11):1018-29. doi: 10.1097/NEN.0b013e318272caab. J Neuropathol Exp Neurol. 2012. PMID: 23095849 Free PMC article. - Small GTPases of the Rab and Arf Families: Key Regulators of Intracellular Trafficking in Neurodegeneration.
Arrazola Sastre A, Luque Montoro M, Lacerda HM, Llavero F, Zugaza JL. Arrazola Sastre A, et al. Int J Mol Sci. 2021 Apr 23;22(9):4425. doi: 10.3390/ijms22094425. Int J Mol Sci. 2021. PMID: 33922618 Free PMC article. Review. - Sorting receptors at Down's syndrome synapses.
Jones MW. Jones MW. Nat Med. 2013 Apr;19(4):404-6. doi: 10.1038/nm.3152. Nat Med. 2013. PMID: 23558625 No abstract available. - Membrane trafficking in neuronal maintenance and degeneration.
Wang D, Chan CC, Cherry S, Hiesinger PR. Wang D, et al. Cell Mol Life Sci. 2013 Aug;70(16):2919-34. doi: 10.1007/s00018-012-1201-4. Epub 2012 Nov 8. Cell Mol Life Sci. 2013. PMID: 23132096 Free PMC article. Review. - Targeted Quantitative Proteomic Approach for High-Throughput Quantitative Profiling of Small GTPases in Brain Tissues of Alzheimer's Disease Patients.
Huang M, Darvas M, Keene CD, Wang Y. Huang M, et al. Anal Chem. 2019 Oct 1;91(19):12307-12314. doi: 10.1021/acs.analchem.9b02485. Epub 2019 Sep 10. Anal Chem. 2019. PMID: 31460748 Free PMC article.
References
- Hyman BT, Van Hoesen GW, Damasio AR, Barnes CL. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;225:1168–1170. - PubMed
- Kordower JH, Chu Y, Stebbins GT, DeKosky ST, Cochran EJ, Bennett D, et al. Loss and atrophy of layer II entorhinal cortex neurons in elderly people with mild cognitive impairment. Ann Neurol. 2001;49:202–213. - PubMed
- Hyman BT, Van Hoesen GW, Damasio AR. Memory-related neural systems in Alzheimer’s disease: an anatomic study. Neurology. 1990;40:1721–1730. - PubMed
- Ginsberg SD, Schmidt ML, Crino PB, Eberwine JH, Lee VM-Y, Trojanowski JQ. Molecular pathology of Alzheimer’s disease and related disorders. In: Peters A, Morrison JH, editors. Cerebral Cortex, vol 14 Neurodegenerative and Age-related Changes in Structure and Function of Cerebral Cortex. New York: Kluwer Academic/Plenum; 1999. pp. 603–653.
Publication types
MeSH terms
Substances
Grants and funding
- AG14449/AG/NIA NIH HHS/United States
- P01 AG014449-12/AG/NIA NIH HHS/United States
- P01 AG009466/AG/NIA NIH HHS/United States
- AG10161/AG/NIA NIH HHS/United States
- P01 AG009466-18/AG/NIA NIH HHS/United States
- P01 AG017617/AG/NIA NIH HHS/United States
- AG17617/AG/NIA NIH HHS/United States
- R01 AG043375/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P01 AG017617-100008/AG/NIA NIH HHS/United States
- P01 AG014449/AG/NIA NIH HHS/United States
- R01 NS043939/NS/NINDS NIH HHS/United States
- AG09466/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous