MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function - PubMed (original) (raw)
Comparative Study
MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function
Lindsay B McKenna et al. Gastroenterology. 2010 Nov.
Abstract
Background & aims: Whereas the importance of microRNA (miRNA) for the development of several tissues is well established, its role in the intestine is unknown. We aimed to quantify the complete miRNA expression profile of the mammalian intestinal mucosa and to determine the contribution of miRNAs to intestinal homeostasis using genetic means.
Methods: We determined the miRNA transcriptome of the mouse intestinal mucosa using ultrahigh throughput sequencing. Using high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP), we identified miRNA-messenger RNA target relationships in the jejunum. We employed gene ablation of the obligatory miRNA-processing enzyme Dicer1 to derive mice deficient for all miRNAs in intestinal epithelia.
Results: miRNA abundance varies dramatically in the intestinal mucosa, from 1 read per million to 250,000. Of the 453 miRNA families identified, mmu-miR-192 is the most highly expressed in both the small and large intestinal mucosa, and there is a 53% overlap in the top 15 expressed miRNAs between the 2 tissues. The intestinal epithelium of Dicer1(loxP/loxP);Villin-Cre mutant mice is disorganized, with a decrease in goblet cells, a dramatic increase in apoptosis in crypts of both jejunum and colon, and accelerated jejunal cell migration. Furthermore, intestinal barrier function is impaired in Dicer1-deficient mice, resulting in intestinal inflammation with lymphocyte and neutrophil infiltration. Our list of miRNA-messenger RNA targeting relationships in the small intestinal mucosa provides insight into the molecular mechanisms behind the phenotype of Dicer1 mutant mice.
Conclusions: We have identified all intestinal miRNAs and shown using gene ablation of Dicer1 that miRNAs play a vital role in the differentiation and function of the intestinal epithelium.
Copyright © 2010 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that no conflict of interest exists.
Figures
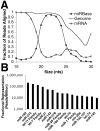
Figure 1. The miRNA transcriptome of murine small intestinal epithelium
(A) Small RNAs between 19 and 25 nucleotides in length, the size of known miRNAs, align to both miRBase and the mouse genome (mm8) but not to mRNA sequences from RefSeq. (B) The fifteen most abundant miRNAs, with relative expression levels expressed as reads per million.
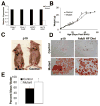
Figure 2. Mice with conditional ablation of Dicer1 in the intestinal epithelium display impaired growth, fat absorption, and water retention
(A) Cre/loxP-mediated gene ablation of Dicer1 was verified by determination of expression levels of Dicer1, mmu-miR-21, and mmu-Let-7b in the intestinal epithelium of control (_Dicer1_loxP/+) and mutant (Dicer1loxP/loxP;Villin-Cre) mice. (B) Dicer1 mutants are growth retarded from ten to fifty days of age, but regain weight thereafter to catch up with controls when fed normal chow (* p < .05). (C) Size comparison of representative pre-weaned (p19) mutant and control littermate. (D) Oil-RedO staining of fecal smears showing fatty stool in mutant pups pre-weaning (p19), and adult mutants on high fat chow as compared to controls. (E) Dicer1 mutants fail to absorb water in the colon, as evidenced by the high water content of their stool compared to control littermates (* p < .05).
Figure 3. Intestine-specific Dicer1 mutants display an expanded crypt zone in the small intestine, an increase in apoptotic cells and a reduction in goblet cell number in the colon
Adult Dicer1 mutants (B) have an expanded crypt zone (noted by *[) in the small intestine (hematoxylin & eosin stained sections) as compared to littermate controls (A). The colonic crypts of adult Dicer1 mutants (D) are disorganized compared to control mice (C). There is in an increase in apoptotic cells, as shown by TUNEL staining, in both the small and large intestine of the mutants (F,H) as compared to controls (E,G). There is also a pronounced decrease in mucus-filled goblet cells in Dicer1 mutants as compared to control in the colon, as shown by Alcian blue staining (I–K) (*** p<.001).
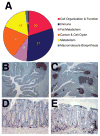
Figure 4. Increased inflammation in the intestine of Dicer1 mutants
(A) Global gene expression profiles of the small intestinal epithelium of Dicer1 mutants and controls were determined using the Agilent Whole Mouse Genome Array. The differentially expressed genes were sorted into KEGG pathways using Gene Set Enrichment Analysis, and the differentially activated pathways combined into functional groups. Immune pathways made up a third of the differentially expressed genes. (B,C) Low magnification (4x) image of large intestine stained for B cells (CD45R) shows drastic increase in lymphoid nodules. (F,G) In the large intestine Dicer1 mutants (G) there is an increase in the number of B cells as compared to controls (F).
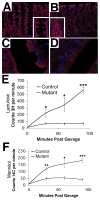
Figure 5. Dicer1 mutants exhibit a disorganized epithelium and decreased tight junctions leading to increased intestinal permeability
(A) Claudin-7 staining shows an extremely disorganized colonic epithelium of Dicer1 mutants (B) as compared to controls (A) insert shows details at higher magnification. (C,D) Claudin-4 staining marks tight junctions in the small intestine. There is a decrease and disorganization of tight junctions in the small intestine of Dicer1 mutants (D) as compared to controls (C). (E,F) (E,F) Dicer1 mutants have increased intestinal permeability as shown by lactulose (E) and mannitol (F) absorption (* p >.05. ** p > .01, *** p>.0001).
Similar articles
- MicroRNAs regulate pituitary development, and microRNA 26b specifically targets lymphoid enhancer factor 1 (Lef-1), which modulates pituitary transcription factor 1 (Pit-1) expression.
Zhang Z, Florez S, Gutierrez-Hartmann A, Martin JF, Amendt BA. Zhang Z, et al. J Biol Chem. 2010 Nov 5;285(45):34718-28. doi: 10.1074/jbc.M110.126441. Epub 2010 Aug 31. J Biol Chem. 2010. PMID: 20807761 Free PMC article. - Hepatic function is preserved in the absence of mature microRNAs.
Hand NJ, Master ZR, Le Lay J, Friedman JR. Hand NJ, et al. Hepatology. 2009 Feb;49(2):618-26. doi: 10.1002/hep.22656. Hepatology. 2009. PMID: 19127519 Free PMC article. - Dicer1-mediated miRNA processing shapes the mRNA profile and function of murine platelets.
Rowley JW, Chappaz S, Corduan A, Chong MM, Campbell R, Khoury A, Manne BK, Wurtzel JG, Michael JV, Goldfinger LE, Mumaw MM, Nieman MT, Kile BT, Provost P, Weyrich AS. Rowley JW, et al. Blood. 2016 Apr 7;127(14):1743-51. doi: 10.1182/blood-2015-07-661371. Epub 2016 Jan 14. Blood. 2016. PMID: 26773046 Free PMC article. - MicroRNAs in oligodendrocyte and Schwann cell differentiation.
Dugas JC, Notterpek L. Dugas JC, et al. Dev Neurosci. 2011;33(1):14-20. doi: 10.1159/000323919. Epub 2011 Feb 23. Dev Neurosci. 2011. PMID: 21346322 Free PMC article. Review. - miRNAs, 'stemness' and skin.
Aberdam D, Candi E, Knight RA, Melino G. Aberdam D, et al. Trends Biochem Sci. 2008 Dec;33(12):583-91. doi: 10.1016/j.tibs.2008.09.002. Epub 2008 Oct 8. Trends Biochem Sci. 2008. PMID: 18848452 Review.
Cited by
- MicroRNA-Mediated Regulation of Initial Host Responses in a Symbiotic Organ.
Moriano-Gutierrez S, Ruby EG, McFall-Ngai MJ. Moriano-Gutierrez S, et al. mSystems. 2021 May 11;6(3):e00081-21. doi: 10.1128/mSystems.00081-21. mSystems. 2021. PMID: 33975964 Free PMC article. - Intestinal Lipid Metabolism Genes Regulated by miRNAs.
Ruiz-Roso MB, Gil-Zamorano J, López de Las Hazas MC, Tomé-Carneiro J, Crespo MC, Latasa MJ, Briand O, Sánchez-López D, Ortiz AI, Visioli F, Martínez JA, Dávalos A. Ruiz-Roso MB, et al. Front Genet. 2020 Jul 10;11:707. doi: 10.3389/fgene.2020.00707. eCollection 2020. Front Genet. 2020. PMID: 32742270 Free PMC article. - MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients.
Capuano M, Iaffaldano L, Tinto N, Montanaro D, Capobianco V, Izzo V, Tucci F, Troncone G, Greco L, Sacchetti L. Capuano M, et al. PLoS One. 2011;6(12):e29094. doi: 10.1371/journal.pone.0029094. Epub 2011 Dec 15. PLoS One. 2011. PMID: 22194996 Free PMC article. - MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions.
Cichon C, Sabharwal H, Rüter C, Schmidt MA. Cichon C, et al. Tissue Barriers. 2014 Aug 8;2(4):e944446. doi: 10.4161/21688362.2014.944446. eCollection 2014. Tissue Barriers. 2014. PMID: 25610754 Free PMC article.
References
- Lee RCAV. An Extensive Class of Small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
- Lewis BPBC, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
- Bartel D. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK050306/DK/NIDDK NIH HHS/United States
- R01 DK053839-14/DK/NIDDK NIH HHS/United States
- R01-DK053839/DK/NIDDK NIH HHS/United States
- T32-HD007516/HD/NICHD NIH HHS/United States
- R01 DK079881/DK/NIDDK NIH HHS/United States
- R01 DK053839/DK/NIDDK NIH HHS/United States
- R37 DK053839/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical