Epigenetic analysis of KSHV latent and lytic genomes - PubMed (original) (raw)
Epigenetic analysis of KSHV latent and lytic genomes
Zsolt Toth et al. PLoS Pathog. 2010.
Abstract
Epigenetic modifications of the herpesviral genome play a key role in the transcriptional control of latent and lytic genes during a productive viral lifecycle. In this study, we describe for the first time a comprehensive genome-wide ChIP-on-Chip analysis of the chromatin associated with the Kaposi's sarcoma-associated herpesvirus (KSHV) genome during latency and lytic reactivation. Depending on the gene expression class, different combinations of activating [acetylated H3 (AcH3) and H3K4me3] and repressive [H3K9me3 and H3K27me3] histone modifications are associated with the viral latent genome, which changes upon reactivation in a manner that is correlated with their expression. Specifically, both the activating marks co-localize on the KSHV latent genome, as do the repressive marks. However, the activating and repressive histone modifications are mutually exclusive of each other on the bulk of the latent KSHV genome. The genomic region encoding the IE genes ORF50 and ORF48 possesses the features of a bivalent chromatin structure characterized by the concomitant presence of the activating H3K4me3 and the repressive H3K27me3 marks during latency, which rapidly changes upon reactivation with increasing AcH3 and H3K4me3 marks and decreasing H3K27me3. Furthermore, EZH2, the H3K27me3 histone methyltransferase of the Polycomb group proteins (PcG), colocalizes with the H3K27me3 mark on the entire KSHV genome during latency, whereas RTA-mediated reactivation induces EZH2 dissociation from the genomic regions encoding IE and E genes concurrent with decreasing H3K27me3 level and increasing IE/E lytic gene expression. Moreover, either the inhibition of EZH2 expression by a small molecule inhibitor DZNep and RNAi knockdown, or the expression of H3K27me3-specific histone demethylases apparently induced the KSHV lytic gene expression cascade. These data indicate that histone modifications associated with the KSHV latent genome are involved in the regulation of latency and ultimately in the control of the temporal and sequential expression of the lytic gene cascade. In addition, the PcG proteins play a critical role in the control of KSHV latency by maintaining a reversible heterochromatin on the KSHV lytic genes. Thus, the regulation of the spatial and temporal association of the PcG proteins with the KSHV genome may be crucial for propagating the KSHV lifecycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
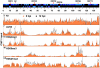
Figure 1. Genome-wide mapping of histone modifications on the KSHV genome during latency and reactivation.
Each ChIP-on-chip experiment is an average of two biological replicates. The histone H3 and histone modification ChIPs were performed with non-induced and doxycycline-induced (12 hpi) TRExBCBL1-Rta cells followed by the hybridization of the labelled ChIP and input DNAs onto a custom designed KSHV-specific 15-bp tiling microarray. See the Material and methods for details. Orange colour indicates 0 hpi-ChIP/input ratio while the black line shows 12 hpi-ChIP/input. Numbers in the left upper corners show the maximum values of Cy5/Cy3. Missing probes in specific genomic regions are shown below the genome scale (**). The alternating dark and light blue squares atop display the viral ORFs where the white triangle indicates ORFs that are expressed from the reverse DNA strand. The “hpi” stands for hours post-induction.
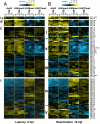
Figure 2. Hierarchical clustering of histone modifications associated with the regulatory regions of viral genes.
Based on their expression patterns the viral genes were grouped as latent (La), IE, E and L genes and hierarchical clustering was performed within the groups. The rows display the histone modification patterns along the −1 kb to +1 kb genomic regions relative to the translational start site (TSS) of each viral gene, which we assigned for the gene regulatory regions. The 1 kb regions are divided into twenty 50 bp fragments that show the average of log2 ratio of probe signal intensities derived from the average of the biological replicates of ChIP-on-chip experiments. Blue and yellow colours represent lower-than-average and higher-than-average for enrichment, respectively, whereas gray shows missing values for enrichment due to lack of probes in those genomic regions. I-V represents the clusters of genes that have similar histone modification patterns. (A) Distinctive histone modification patterns are associated with the KSHV genes of different expression classes during latency. (B) Changes in the enrichment of histone modifications during reactivation (12hpi).
Figure 3. Dynamic association of histone modifications with viral genes during latency and reactivation.
(A) Time-course ChIP analysis of histone modifications on the RTA promoter during latency and reactivation. Cellular controls can be seen in panel D. (B) Colocalization of H3K4me3 and H3K27me3 on the RTA promoter is confirmed by sequential ChIP assays. The first ChIP was performed with either H3K4me3-specific or H3K27me3-specific antibody, followed by the elution of the immunoprecipitated DNAs and a second ChIP with either H3K27me3 or H3K4me3 antibody. LANA and ORF25 promoters were used as controls. (C) Time-course ChIP analysis of histone modifications on the selected latent (LANA), early (K2, ORF56) and late (ORFs 8, 25, 64) genes. Cellular controls can be found in panel D. Pr: promoter, in: within gene body. (D) ChIP assays of histone modifications on cellular promoters. The promoters of the repressed cellular MYT1 and HTF6 genes as well as the active promoter of the actin (ACT) gene were also tested using the same ChIP samples that had been used in panels A and C. ND: not detectable.
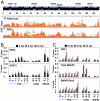
Figure 4. Genome-wide binding of EZH2 to the KSHV genome correlates with the repression of lytic genes.
(A) ChIP-on-chip was performed for EZH2 and its genome-wide binding was compared with the distribution of H3K27me3 on the KSHV genome during both latency and reactivation. Labels are the same as in Figure 1. The H3K27me3 graph was taken from Figure 1. (B) EZH2 and SUZ12 binding to the H3K27me3-rich lytic promoters are shown by independent ChIP assays. EZH2-interacting PcG protein SUZ12 is enriched only where EZH2 is present. (C) Recruitment of transcription activators to the activated lytic promoters during KSHV reactivation. An anti-RNA polymerase II antibody (H-224) that recognizes the RNAPII independently from its phosphorylation state (total RNAPII) was used for ChIP of total RNAPII, while the anti-RNA polymerase II antibody CTD4H8 specifically immunoprecipitates RNAPII phosphorylated at the 5th serine of its C-terminal domain (RNAPII Ser5).
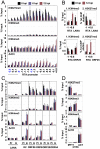
Figure 5. Polycomb group proteins are involved in the maintenance of latency of KSHV.
(A) Overexpression of the wild-type HA-tagged H3K27me3 histone methyltransferases (HMTs) UTX and JMJD3 triggers the lytic reactivation of KSHV in Vero-rKSHV.219 as shown with the expression of RFP. In contrast, the H3K9me3 HMT JMJD2A and the enzymatically inactive UTXm showed little or no effect on KSHV lytic reactivation. (B) Quantification of RFP positive cells. (C and D) JSC-1 cells were treated with 5 uM DZNep for 1, 2 and 3 days and the cells were harvested for immunoblot analysis with the indicated specific antibodies against cellular proteins and histone modifications (C) or viral proteins (D). “Dpt” indicates days post-treatment. Whole cell lysate of NaB-treated JSC-1 cells was used as a control for Zta immunoblot. (E) JSC-1 cells were treated with DZNep as described in (C) and total RNAs were isolated for RT-qPCR analysis of some selected KSHV, EBV and cellular mRNAs. (F and G) BCBL-1 cells were infected by lentivirus expressing the indicated shRNAs and were then subject to immunoblotting analysis with the indicated antibodies (F) or RT-qPCR analysis was performed for the indicated viral transcripts (G).
Similar articles
- Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection.
Toth Z, Brulois K, Lee HR, Izumiya Y, Tepper C, Kung HJ, Jung JU. Toth Z, et al. PLoS Pathog. 2013;9(12):e1003813. doi: 10.1371/journal.ppat.1003813. Epub 2013 Dec 19. PLoS Pathog. 2013. PMID: 24367262 Free PMC article. - ZIC2 Is Essential for Maintenance of Latency and Is a Target of an Immediate Early Protein during Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation.
Lyu Y, Nakano K, Davis RR, Tepper CG, Campbell M, Izumiya Y. Lyu Y, et al. J Virol. 2017 Oct 13;91(21):e00980-17. doi: 10.1128/JVI.00980-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28835494 Free PMC article. - Molecular biology of KSHV lytic reactivation.
Purushothaman P, Uppal T, Verma SC. Purushothaman P, et al. Viruses. 2015 Jan 14;7(1):116-53. doi: 10.3390/v7010116. Viruses. 2015. PMID: 25594835 Free PMC article. Review. - Rewriting Viral Fate: Epigenetic and Transcriptional Dynamics in KSHV Infection.
Han C, Niu D, Lan K. Han C, et al. Viruses. 2024 Nov 30;16(12):1870. doi: 10.3390/v16121870. Viruses. 2024. PMID: 39772181 Free PMC article. Review.
Cited by
- A cultured affair: HSV latency and reactivation in neurons.
Wilson AC, Mohr I. Wilson AC, et al. Trends Microbiol. 2012 Dec;20(12):604-11. doi: 10.1016/j.tim.2012.08.005. Epub 2012 Sep 7. Trends Microbiol. 2012. PMID: 22963857 Free PMC article. Review. - Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi's sarcoma-associated herpesvirus lytic gene expression.
Toth Z, Brulois KF, Wong LY, Lee HR, Chung B, Jung JU. Toth Z, et al. J Virol. 2012 Sep;86(18):9696-707. doi: 10.1128/JVI.01012-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740393 Free PMC article. - LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection.
Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU. Toth Z, et al. PLoS Pathog. 2016 Sep 8;12(9):e1005878. doi: 10.1371/journal.ppat.1005878. eCollection 2016 Sep. PLoS Pathog. 2016. PMID: 27606464 Free PMC article. - Transcriptional and post-transcriptional regulation of viral gene expression in the gamma-herpesvirus Kaposi's sarcoma-associated herpesvirus.
Butnaru M, Gaglia MM. Butnaru M, et al. Curr Clin Microbiol Rep. 2018 Dec;5(4):219-228. doi: 10.1007/s40588-018-0102-1. Epub 2018 Aug 3. Curr Clin Microbiol Rep. 2018. PMID: 30854283 Free PMC article. - Epigenetic control in Kaposi sarcoma-associated herpesvirus infection and associated disease.
Fröhlich J, Grundhoff A. Fröhlich J, et al. Semin Immunopathol. 2020 Apr;42(2):143-157. doi: 10.1007/s00281-020-00787-z. Epub 2020 Mar 26. Semin Immunopathol. 2020. PMID: 32219477 Free PMC article. Review.
References
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- MacDonald VE, Howe LJ. Histone acetylation: where to go and how to get there. Epigenetics. 2009;4:139–143. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 DE019085/DE/NIDCR NIH HHS/United States
- AI073099/AI/NIAID NIH HHS/United States
- R01 CA082057/CA/NCI NIH HHS/United States
- R01 CA115284/CA/NCI NIH HHS/United States
- R01 AI073099/AI/NIAID NIH HHS/United States
- RC2 CA148616/CA/NCI NIH HHS/United States
- R01 CA031363/CA/NCI NIH HHS/United States
- CA148616/CA/NCI NIH HHS/United States
- DE019085/DE/NIDCR NIH HHS/United States
- CA31363/CA/NCI NIH HHS/United States
- CA082057/CA/NCI NIH HHS/United States
- CA115284/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources