A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana - PubMed (original) (raw)
A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana
Marco Todesco et al. PLoS Genet. 2010.
Abstract
Many targets of plant microRNAs (miRNAs) are thought to play important roles in plant physiology and development. However, because plant miRNAs are typically encoded by medium-size gene families, it has often been difficult to assess their precise function. We report the generation of a large-scale collection of knockdowns for Arabidopsis thaliana miRNA families; this has been achieved using artificial miRNA target mimics, a recently developed technique fashioned on an endogenous mechanism of miRNA regulation. Morphological defects in the aerial part were observed for approximately 20% of analyzed families, all of which are deeply conserved in land plants. In addition, we find that non-cleavable mimic sites can confer translational regulation in cis. Phenotypes of plants expressing target mimics directed against miRNAs involved in development were in several cases consistent with previous reports on plants expressing miRNA-resistant forms of individual target genes, indicating that a limited number of targets mediates most effects of these miRNAs. That less conserved miRNAs rarely had obvious effects on plant morphology suggests that most of them do not affect fundamental aspects of development. In addition to insight into modes of miRNA action, this study provides an important resource for the study of miRNA function in plants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
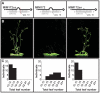
Figure 1. Requirement of a bulge at the cleavage site for target mimicry.
(A) A target mimic with an unmodified central sequence (MIM172cs), which retained complementarity to the central portion of miR172 across the cleavage site (red line) opposite position 10 to 11 of the miRNA, did not change flowering time. Modification of the central sequence (TCTA to GAGT; MIM172) restored a three nucleotide bulge found in IPS1 and generated a functional target mimic, causing a delay in flowering. However, a single nucleotide mismatch introduced into the center of an authentic miR172 target site (MIM172sn), but without a bulge, was not sufficient to reduce miR172 activity. (B) Four-week old plants grown at 23°C in long days. MIM172cs and MIM172sn are phenotypically indistinguishable from wild-type Col-0 plants (see also Figure S1B). (C) Distribution of flowering times of primary transformants grown in the same conditions; compare with Col-0 plants transformed with an empty binary vector in Figure S2.
Figure 2. Leaf rosettes of target mimic expressing plants.
Three-week-old plants. Bar corresponds to 1 cm for all panels.
Figure 3. Details of defects observed in target mimic expressing plants.
(A) Smaller flowers in severe MIM319 lines. The most strongly affected flowers lacked petals and did not have fully developed anthers (left side); in milder lines, flowers had short sepals, narrow petals, but were fertile (middle). Two flowers from wild type Col-0 are shown on the right side of the panel. (B) Severe MIM159 and MIM319 lines were very small and compact, without any stem elongation. (C) Leaves of MIM164 plants (compared to a leaf from wild type Col-0, on the far left). (D, E) Developing fruits of MIM164 with ectopic growths emanating from valve margins (D), which can develop into pseudo-pistils in severe lines (E). (F) Anthers in MIM167 lines did not mature completely (top), resulting in reduced pollen production (compared to a wild type Col-0 flower, bottom). (G) Seeds of MIM167 plants often do not fill completely, and remained attached to the dried silique (compared to a silique of wild type Col-0, on the right). (H, I) MIM171A lines suffered from defects in the separation of sepals, which prevented emergence of the pistil (H), and caused the plants to be mostly sterile (I, on the left, compared to a wild-type Col-0 plant, on the right). Bars correspond to 1 cm in (A–C) and I, and to 0.1 cm in (D–H).
Figure 4. Effects of artificial mimics on levels of miRNAs and miRNA targets.
(A) Nine-day-old plants. Introduction of a MIM159 fragment into the 3′ UTR silences a constitutively expressed 3xEYFP in the MIR159 expression domain (compare p35S:3xEYFP and p35S:3xEYFP-MIM159), which is revealed in the pMIR159:GUS lines. MiR159 activity is also indirectly revealed by comparing the effect of expressing MIM159 in a genomic MYB33:GUS line. (B) Transcript levels of select miRNA targets in two independent lines for each MIM construct (represented by bars of different shades of gray). (C) Expression levels of miRNA targets in mutants impaired in miRNA biogenesis or targeting. Expression values are reported as the average of two biological and two technical replicates, and are normalized to the expression levels in wild type Col-0 plants (dotted line). (D) CIP4 mRNA and protein levels in four independent MIM834 lines. Band intensity relative to the wild-type control is reported. (E) Levels of mature miRNAs in several MIM lines. U6 accumulation is shown as control. Increased accumulation of miR156 (lower band in the blot) was observed upon expression of a resistant version of a miR156 target (consistent with what observed for miRNA156a precursor levels in [39]) or inhibition of miRNA activity in the _ago1_-27 mutants. The decrease in miR156 levels in MIM156 plants is then not an indirect consequence of increased SPL transcript levels.
Similar articles
- The Use of MicroRNA Decoy Technologies to Inhibit miRNA Function in Arabidopsis.
Wong G, Millar AA. Wong G, et al. Methods Mol Biol. 2019;1932:227-238. doi: 10.1007/978-1-4939-9042-9_17. Methods Mol Biol. 2019. PMID: 30701504 - MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana.
Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, Carrington JC. Fahlgren N, et al. Plant Cell. 2010 Apr;22(4):1074-89. doi: 10.1105/tpc.110.073999. Epub 2010 Apr 20. Plant Cell. 2010. PMID: 20407027 Free PMC article. - A virus-based miRNA suppression (VbMS) system for miRNA loss-of-function analysis in plants.
Yan F, Guo W, Wu G, Lu Y, Peng J, Zheng H, Lin L, Chen J. Yan F, et al. Biotechnol J. 2014 May;9(5):702-8. doi: 10.1002/biot.201300523. Epub 2014 Apr 10. Biotechnol J. 2014. PMID: 24664983 - Alternate approaches to repress endogenous microRNA activity in Arabidopsis thaliana.
Eamens AL, Wang MB. Eamens AL, et al. Plant Signal Behav. 2011 Mar;6(3):349-59. doi: 10.4161/psb.6.3.14340. Epub 2011 Mar 1. Plant Signal Behav. 2011. PMID: 21358288 Free PMC article. Review. - Overview of Repressive miRNA Regulation by Short Tandem Target Mimic (STTM): Applications and Impact on Plant Biology.
Othman SMIS, Mustaffa AF, Che-Othman MH, Samad AFA, Goh HH, Zainal Z, Ismail I. Othman SMIS, et al. Plants (Basel). 2023 Feb 3;12(3):669. doi: 10.3390/plants12030669. Plants (Basel). 2023. PMID: 36771753 Free PMC article. Review.
Cited by
- Identification and Characterization of MiRNAs in Coccomyxa subellipsoidea C-169.
Yang R, Chen G, Peng H, Wei D. Yang R, et al. Int J Mol Sci. 2019 Jul 13;20(14):3448. doi: 10.3390/ijms20143448. Int J Mol Sci. 2019. PMID: 31337051 Free PMC article. - Contribution of Omics and Systems Biology to Plant Biotechnology.
Dalio RJD, Litholdo CG Jr, Arena G, Magalhães D, Machado MA. Dalio RJD, et al. Adv Exp Med Biol. 2021;1346:171-188. doi: 10.1007/978-3-030-80352-0_10. Adv Exp Med Biol. 2021. PMID: 35113402 Review. - MicroRNA activity in the Arabidopsis male germline.
Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD. Borges F, et al. J Exp Bot. 2011 Mar;62(5):1611-20. doi: 10.1093/jxb/erq452. J Exp Bot. 2011. PMID: 21357774 Free PMC article. Review. - A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana.
Wang L, Zhou CM, Mai YX, Li LZ, Gao J, Shang GD, Lian H, Han L, Zhang TQ, Tang HB, Ren H, Wang FX, Wu LY, Liu XL, Wang CS, Chen EW, Zhang XN, Liu C, Wang JW. Wang L, et al. EMBO J. 2019 Apr 15;38(8):e100063. doi: 10.15252/embj.2018100063. Epub 2019 Mar 6. EMBO J. 2019. PMID: 30842098 Free PMC article. - Regulation of gene expression in plants through miRNA inactivation.
Ivashuta S, Banks IR, Wiggins BE, Zhang Y, Ziegler TE, Roberts JK, Heck GR. Ivashuta S, et al. PLoS One. 2011;6(6):e21330. doi: 10.1371/journal.pone.0021330. Epub 2011 Jun 23. PLoS One. 2011. PMID: 21731706 Free PMC article.
References
- Dugas DV, Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol. 2008;67:403–417. - PubMed
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources