Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development - PubMed (original) (raw)
Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development
Yongqun He et al. J Biomed Biotechnol. 2010.
Abstract
Vaxign is the first web-based vaccine design system that predicts vaccine targets based on genome sequences using the strategy of reverse vaccinology. Predicted features in the Vaxign pipeline include protein subcellular location, transmembrane helices, adhesin probability, conservation to human and/or mouse proteins, sequence exclusion from genome(s) of nonpathogenic strain(s), and epitope binding to MHC class I and class II. The precomputed Vaxign database contains prediction of vaccine targets for >70 genomes. Vaxign also performs dynamic vaccine target prediction based on input sequences. To demonstrate the utility of this program, the vaccine candidates against uropathogenic Escherichia coli (UPEC) were predicted using Vaxign and compared with various experimental studies. Our results indicate that Vaxign is an accurate and efficient vaccine design program.
Figures
Figure 1
The Vaxign algorithm pipeline.
Figure 2
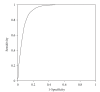
ROC curve analysis of epitopes binding HLA A*0201.
Figure 3
Vaxign prediction of UPEC vaccine targets. (a) Selection of E. coli CFT073 as a seed genome. Different filter options are applied: only outer membrane proteins, maximum of one transmembrane helix, minimum adhesin probability of.51, having orthologs in other three UPEC genomes but no orthologs in K-12 strain M1655, and no similarity to any human or mouse proteins. (b) In total 22 proteins were predicted to meet selected criteria. (c) Detailed results for a particular protein, for example, NmpC. MHC Class I and II epitope prediction results are also available and can be further filtered. (d) Prediction of _α_-helices and their possible cellular locations of NmpC. (e) Prediction of _β_-barrels and their possible cellular locations of NmpC.
Figure 4
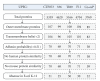
Prediction of UPEC vaccine targets conserved in four sequenced UPEC genomes using Vaxign. Note: * Co-ed represents the conserved proteins.
Similar articles
- Vaxign2: the second generation of the first Web-based vaccine design program using reverse vaccinology and machine learning.
Ong E, Cooke MF, Huffman A, Xiang Z, Wong MU, Wang H, Seetharaman M, Valdez N, He Y. Ong E, et al. Nucleic Acids Res. 2021 Jul 2;49(W1):W671-W678. doi: 10.1093/nar/gkab279. Nucleic Acids Res. 2021. PMID: 34009334 Free PMC article. - Genome-wide prediction of vaccine targets for human herpes simplex viruses using Vaxign reverse vaccinology.
Xiang Z, He Y. Xiang Z, et al. BMC Bioinformatics. 2013;14 Suppl 4(Suppl 4):S2. doi: 10.1186/1471-2105-14-S4-S2. Epub 2013 Mar 8. BMC Bioinformatics. 2013. PMID: 23514126 Free PMC article. - Vaccine Design by Reverse Vaccinology and Machine Learning.
Ong E, He Y. Ong E, et al. Methods Mol Biol. 2022;2414:1-16. doi: 10.1007/978-1-0716-1900-1_1. Methods Mol Biol. 2022. PMID: 34784028 - Databases and in silico tools for vaccine design.
He Y, Xiang Z. He Y, et al. Methods Mol Biol. 2013;993:115-27. doi: 10.1007/978-1-62703-342-8_8. Methods Mol Biol. 2013. PMID: 23568467 Review. - Comparison of Open-Source Reverse Vaccinology Programs for Bacterial Vaccine Antigen Discovery.
Dalsass M, Brozzi A, Medini D, Rappuoli R. Dalsass M, et al. Front Immunol. 2019 Feb 14;10:113. doi: 10.3389/fimmu.2019.00113. eCollection 2019. Front Immunol. 2019. PMID: 30837982 Free PMC article. Review.
Cited by
- Immunoinformatics Design of a Multiepitope Vaccine (MEV) Targeting Streptococcus mutans: A Novel Computational Approach.
Naorem RS, Pangabam BD, Bora SS, Fekete C, Teli AB. Naorem RS, et al. Pathogens. 2024 Oct 21;13(10):916. doi: 10.3390/pathogens13100916. Pathogens. 2024. PMID: 39452787 Free PMC article. - Enhancing Vaxign-DL for Vaccine Candidate Prediction with added ESM-Generated Features.
Chen Y, Zhang Y, He Y. Chen Y, et al. bioRxiv [Preprint]. 2024 Sep 8:2024.09.04.611295. doi: 10.1101/2024.09.04.611295. bioRxiv. 2024. PMID: 39282385 Free PMC article. Preprint. - Advanced vaccinomic, immunoinformatic, and molecular modeling strategies for designing Multi- epitope vaccines against the Enterobacter cloacae complex.
Alhassan HH. Alhassan HH. Front Immunol. 2024 Aug 16;15:1454394. doi: 10.3389/fimmu.2024.1454394. eCollection 2024. Front Immunol. 2024. PMID: 39221241 Free PMC article. - In silico and in vivo Investigations of the Immunoreactivity of Klebsiella pneumoniae OmpA Protein as a Vaccine Candidate.
Shahbazi S, Badmasti F, Habibi M, Sabzi S, Noori Goodarzi N, Farokhi M, Asadi Karam MR. Shahbazi S, et al. Iran Biomed J. 2024 Jul 1;28(4):156-67. doi: 10.61186/ibj.4023. Iran Biomed J. 2024. PMID: 38946021 Free PMC article. - From proteome to candidate vaccines: target discovery and molecular dynamics-guided multi-epitope vaccine engineering against kissing bug.
Albaqami FF, Altharawi A, Althurwi HN, Alharthy KM. Albaqami FF, et al. Front Immunol. 2024 Jun 10;15:1413893. doi: 10.3389/fimmu.2024.1413893. eCollection 2024. Front Immunol. 2024. PMID: 38915396 Free PMC article.
References
- Rappuoli R. Reverse vaccinology. Current Opinion in Microbiology. 2000;3(5):445–450. - PubMed
- Pizza M, Scarlato V, Masignani V, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287(5459):1816–1820. - PubMed
- Ross BC, Czajkowski L, Hocking D, et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. 2001;19(30):4135–4142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI081062/AI/NIAID NIH HHS/United States
- AI43363/AI/NIAID NIH HHS/United States
- R01 AI081062-01A1/AI/NIAID NIH HHS/United States
- R56 AI043363/AI/NIAID NIH HHS/United States
- R01 AI043363/AI/NIAID NIH HHS/United States
- R01 AI081062/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials