Our unindicted coconspirators: human metabolism from a microbial perspective - PubMed (original) (raw)
Our unindicted coconspirators: human metabolism from a microbial perspective
Andrew L Goodman et al. Cell Metab. 2010.
Abstract
The human gut is populated by a microbiota composed of tens of trillions of organisms and millions of genes that together form a metabolic "organ." Intra- and interpersonal differences in the structure and functions of this organ suggest that it could contribute to our normal metabolic variations and also to metabolic disorders. This Commentary discusses experimental approaches for connecting microbial and host metabolism in the context of health and disease.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
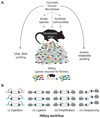
Figure 1. Connecting microbiota and metabolism through gnotobiotics
(A) A complete human gut (fecal) microbiota, obtained from individuals varying in their diet or physiology, can be surveyed by highly parallel DNA-, RNA-, protein-, and metabolite-directed methods. These microbial communities also serve as source material for transplantation into germ-free mice. They can also be fractionated into component species and assembled into “synthetic communities” of defined composition in order to directly test the roles of specific phylotypes in shaping host physiology: these phylotypes can be intentionally added, removed, or substituted with mutagenized populations prior to colonization of germ-free animals to identify genes required for fitness in specific host/dietary contexts using a method known as insertion sequencing (INSeq). (B) Quantifying the relative abundance of tens of thousands of transposon mutants of a given microbial species by INSeq. (i) The INSeq transposon (depicted as an open rectangle) contains recognition sites for the Type IIs restriction enzyme MmeI in its inverted repeats (black triangles); MmeI digests adjacent chromosomal DNA 16bp outside of the transposon. (ii) Following digestion, sequencing adapters (grey ovals) are appended by ligation. These adapters can contain sample-specific barcodes to allow sample pooling. (iii) A limited number of cycles of PCR amplification of these uniformly-sized molecules are performed. (iv) The resulting amplicons are sequenced using a massively parallel sequencing instrument. The sequence of each read indicates the genomic location of the source transposon, and the relative abundance of each sequence mirrors the relative abundance of the corresponding transposon mutant in the population. Comparison of these relative abundances in input versus output microbial populations identifies genes whose functions are required for fitness in vivo.
References
MeSH terms
Grants and funding
- F32 AI078628/AI/NIAID NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- R01 DK070977/DK/NIDDK NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources