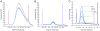Unlocking short read sequencing for metagenomics - PubMed (original) (raw)
Unlocking short read sequencing for metagenomics
Sébastien Rodrigue et al. PLoS One. 2010.
Abstract
Background: Different high-throughput nucleic acid sequencing platforms are currently available but a trade-off currently exists between the cost and number of reads that can be generated versus the read length that can be achieved.
Methodology/principal findings: We describe an experimental and computational pipeline yielding millions of reads that can exceed 200 bp with quality scores approaching that of traditional Sanger sequencing. The method combines an automatable gel-less library construction step with paired-end sequencing on a short-read instrument. With appropriately sized library inserts, mate-pair sequences can overlap, and we describe the SHERA software package that joins them to form a longer composite read.
Conclusions/significance: This strategy is broadly applicable to sequencing applications that benefit from low-cost high-throughput sequencing, but require longer read lengths. We demonstrate that our approach enables metagenomic analyses using the Illumina Genome Analyzer, with low error rates, and at a fraction of the cost of pyrosequencing.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
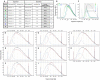
Figure 1. Size-dependent isolation of DNA fragments from sheared genomic DNA via dSPRI.
AMPure XP SPRI beads bind DNA fragments in a size dependent manner according to the concentration of salts and polyethylene glycol (PEG) in the reaction –, which can easily be changed by using different volume ratios of DNA to SPRI bead solutions. A two-step procedure is employed to isolate targeted DNA size fractions. Panels A to H present Bioanalyzer DNA-1000 assays showing the sheared genomic DNA used as starting material (black), the larger size DNA fragments discarded in separation 1 (red), and the size fraction purified and recovered after separation 2 (blue). Panel I is a table summarizing the conditions and results displayed in panels A to H. All Bioanalyzer DNA-1000 traces after separation 1 (panel J), and after separation 2 (panel K), are respectively displayed on a graph for the conditions presented in panels A to H. The conditions displayed in panel H were used to obtaine the Illumina composite reads discussed in the text. The wider DNA fragment size distribution from panel H allowed to better analyze the effects of shorter versus longer overlapping regions on consensus reads.
Figure 2. Reproducibility of double-SPRI.
The panel shows DNA fragment size distributions as obtained by Bioanalyzer DNA-1000 assays. The curves represent the size fractions removed during the first separation step A) or recovered after the second separation B). The two size fractions were independently reproduced in 4 or 8 separation experiments. The curves in B) represent the libraries sequenced after dSPRI based size selection, adapter ligation and PCR enrichment. While concentrations (arbitrary fluorescence units) vary between reproduced libraries the range of removed or enriched DNA fragment sizes was highly reproducible. Panel c) shows the DNA fragment size distribution recovered after the second separation when using decreasing amounts sheared genomic DNA. dSPRI allows reliable size selection in a DNA concentration independent manner.
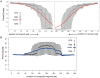
Figure 3. Quality of Illumina reads out to 143 bp.
A) The mean Phred quality of single reads is shown in the solid red lines, with error bars displaying quartiles. Read quality is highly variable toward the reads' ends. Read quality as a function of base pair is worse on the second mate of the pair. B) The mean and quartiles of Phred quality by base for the average-length composite read.
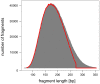
Figure 4. High-confidence alignment yield by insert lengths.
Distribution of insert lengths by aligning the original reads to the reference sequence (gray), and lengths of composite reads retained (red).
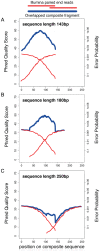
Figure 5. Quality of composite fragments after overlapping.
The mean Phred quality (and corresponding error rate) at each position of the read. We show the original Illumina reads and the composite fragments generated by overlapping. A) 143 bp, the length of the original Illumina read; B) 180 bp, the mean length of overlapped fragment in this library and C) 250 bp, roughly the mean length generated by 454-FLX technology.
Figure 6. Composite Illumina reads constitute a legitimate alternative to pyrosequencing for metagenomics studies.
DNA from a marine metagenomics sample was sequenced with our overlapping mate-pair approach. The composite reads were directly compared to 454-FLX sequences from the exact same sample. A) Fraction of reads that could be assigned to a taxon using the composite reads (mean read length 180 bp), the entire 454-FLX dataset (mean read length 207 bp), or longer 454-FLX reads (mean read length 254 bp). B) Comparison of taxon assignments using composite reads and 454-FLX pyrosequencing reads. The top 25 represented taxa, with colored symbols next to the bracket, are listed in Table S1.
Figure 7. Short insertions and deletions in low-confidence composite Illumina reads.
Mate-reads from the control lane (PhiX174 bacteriophage genome) were used to assess false alignments introduced by the SHERA pipeline. After constructing the composite sequences and filtering, we plotted the difference in length (if any) between a composite fragment and its insert length as predicted by MAQ by mapping the original mate-pairs to the reference.This histogram is a sum of two distributions, the overlapper software's misalignments (a broad gaussian) and a sharp peak of small (1–2 bp) indels. We used a simple and conservative linear model to remove the indels and infer the number of misalignments.
Similar articles
- Short clones or long clones? A simulation study on the use of paired reads in metagenomics.
Mitra S, Schubach M, Huson DH. Mitra S, et al. BMC Bioinformatics. 2010 Jan 18;11 Suppl 1(Suppl 1):S12. doi: 10.1186/1471-2105-11-S1-S12. BMC Bioinformatics. 2010. PMID: 20122183 Free PMC article. - Improving the sensitivity of long read overlap detection using grouped short k-mer matches.
Du N, Chen J, Sun Y. Du N, et al. BMC Genomics. 2019 Apr 4;20(Suppl 2):190. doi: 10.1186/s12864-019-5475-x. BMC Genomics. 2019. PMID: 30967123 Free PMC article. - Pseudo-Sanger sequencing: massively parallel production of long and near error-free reads using NGS technology.
Ruan J, Jiang L, Chong Z, Gong Q, Li H, Li C, Tao Y, Zheng C, Zhai W, Turissini D, Cannon CH, Lu X, Wu CI. Ruan J, et al. BMC Genomics. 2013 Oct 17;14(1):711. doi: 10.1186/1471-2164-14-711. BMC Genomics. 2013. PMID: 24134808 Free PMC article. - Sequence assembly using next generation sequencing data--challenges and solutions.
Chin FY, Leung HC, Yiu SM. Chin FY, et al. Sci China Life Sci. 2014 Nov;57(11):1140-8. doi: 10.1007/s11427-014-4752-9. Epub 2014 Oct 17. Sci China Life Sci. 2014. PMID: 25326069 Review. - Sequencing and genome assembly using next-generation technologies.
Nagarajan N, Pop M. Nagarajan N, et al. Methods Mol Biol. 2010;673:1-17. doi: 10.1007/978-1-60761-842-3_1. Methods Mol Biol. 2010. PMID: 20835789 Review.
Cited by
- Impact of host genetics on gut microbiome: Take-home lessons from human and mouse studies.
Cahana I, Iraqi FA. Cahana I, et al. Animal Model Exp Med. 2020 Sep 17;3(3):229-236. doi: 10.1002/ame2.12134. eCollection 2020 Sep. Animal Model Exp Med. 2020. PMID: 33024944 Free PMC article. Review. - The histone variant H2A.Z is an important regulator of enhancer activity.
Brunelle M, Nordell Markovits A, Rodrigue S, Lupien M, Jacques PÉ, Gévry N. Brunelle M, et al. Nucleic Acids Res. 2015 Nov 16;43(20):9742-56. doi: 10.1093/nar/gkv825. Epub 2015 Aug 28. Nucleic Acids Res. 2015. PMID: 26319018 Free PMC article. - Virioplankton Assemblage Structure in the Lower River and Ocean Continuum of the Amazon.
Silva BSO, Coutinho FH, Gregoracci GB, Leomil L, de Oliveira LS, Fróes A, Tschoeke D, Soares AC, Cabral AS, Ward ND, Richey JE, Krusche AV, Yager PL, de Rezende CE, Thompson CC, Thompson FL. Silva BSO, et al. mSphere. 2017 Oct 4;2(5):e00366-17. doi: 10.1128/mSphere.00366-17. eCollection 2017 Sep-Oct. mSphere. 2017. PMID: 28989970 Free PMC article. - A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps.
Tan SJ, Phan H, Gerry BM, Kuhn A, Hong LZ, Min Ong Y, Poon PS, Unger MA, Jones RC, Quake SR, Burkholder WF. Tan SJ, et al. PLoS One. 2013 Jul 24;8(7):e64084. doi: 10.1371/journal.pone.0064084. Print 2013. PLoS One. 2013. PMID: 23894273 Free PMC article. - The Western English Channel contains a persistent microbial seed bank.
Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. Caporaso JG, et al. ISME J. 2012 Jun;6(6):1089-93. doi: 10.1038/ismej.2011.162. Epub 2011 Nov 10. ISME J. 2012. PMID: 22071345 Free PMC article.
References
- Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–18. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources