Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity - PubMed (original) (raw)
Comparative Study
Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity
Olga T Hardy et al. Surg Obes Relat Dis. 2011 Jan-Feb.
Abstract
Background: Obesity is a strong risk factor for resistance to insulin-mediated glucose disposal, a precursor of type 2 diabetes and other disorders. However, not all obese individuals are insulin resistant. We sought to identify the molecular pathways that might cause obesity-associated insulin resistance in humans by studying the morbidly obese who were insulin sensitive versus insulin resistant, thereby eliminating obesity as a variable.
Methods: Combining gene expression profiling with computational approaches, we determined the global gene expression signatures of omental and subcutaneous adipose tissue samples obtained from similarly obese patients undergoing gastric bypass surgery.
Results: Gene sets related to chemokine activity and chemokine receptor binding were identified as most highly expressed in the omental tissue from insulin-resistant compared with insulin-sensitive subjects, independent of the body mass index. These upregulated genes included chemokines (C-C motif) ligand 2, 3, 4, and 18 and interleukin-8/(CC-X motif) ligand 8 and were not differentially expressed in the subcutaneous adipose tissues between the 2 groups of subjects. Insulin resistance, but not the body mass index, was associated with increased macrophage infiltration in the omental adipose tissue, as was adipocyte size, in these morbidly obese subjects.
Conclusion: Our findings have demonstrated that inflammation of the omental adipose tissue is strongly associated with insulin resistance in human obesity even in subjects with similar body mass index values.
Copyright © 2011 American Society for Metabolic and Bariatric Surgery. Published by Elsevier Inc. All rights reserved.
Figures
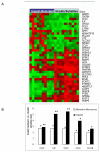
Figure 1
Expression of inflammation-related genes in omental adipose tissue from insulin resistant and insulin sensitive obese human subjects using microarray and RTqPCR. A. Heatmap representing normalized expression of genes identified by Microarray Computational Environment (MACE) as being significantly increased in omental adipose tissue from insulin resistant subjects and insulin sensitive human subjects. The genes are listed on the right. Expression levels above the mean for the gene are shown in red and expression levels below the mean for the gene are shown in green. B. Fold change in mRNA level of genes in omental adipose tissue of obese, insulin-resistant subjects (n=10) relative to obese, insulin-sensitive subjects (n=10) based on microarray data (white bars) and quantitative real-time analysis (black bars) performed by RTqPCR. ** P < 0.01; * P < 0.05.
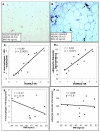
Figure 2
Relationship between macrophage infiltration, adipocyte diameter and HOMA2-IR in omental adipose tissue. Immunohistochemical detection of CD68+ macrophages performed on omental adipose tissue samples obtained from obese human subjects undergoing gastric bypass surgery. A-B. CD68 staining of omental adipose tissue from a representative insulin sensitive (A) and insulin resistant (B) subject. Unlike in adipose tissue from insulin sensitive subject (A) macrophages are observed throughout the tissue (arrows) and arranged in crown-like structures (arrowhead) in the insulin resistant subject (B). Sections taken at 20x magnification. CF, Quantitative analysis of adipocyte diameter and macrophage infiltration in adipose tissue of insulin resistant and insulin sensitive obese human subjects. Adipocyte diameter was calculated from the perimeter measurement of 100 cells. Insulin sensitivity, as determined by HOMA2-IR, correlates with CD68+ macrophage infiltration (C) and adipocyte diameter (D). Body mass index (BMI) shows no correlation with macrophage infiltration (E) or adipocyte diameter (F). Data is an average of 10 histological fields using 10X objective (C or E).
Similar articles
- Omental gene expression of adiponectin correlates with degree of insulin sensitivity before and after gastric bypass surgery.
Chen J, Spagnoli A, Torquati A. Chen J, et al. Obes Surg. 2012 Mar;22(3):472-7. doi: 10.1007/s11695-011-0568-x. Obes Surg. 2012. PMID: 22161113 Free PMC article. - Glucose uptake and insulin action in human adipose tissue--influence of BMI, anatomical depot and body fat distribution.
Stolic M, Russell A, Hutley L, Fielding G, Hay J, MacDonald G, Whitehead J, Prins J. Stolic M, et al. Int J Obes Relat Metab Disord. 2002 Jan;26(1):17-23. doi: 10.1038/sj.ijo.0801850. Int J Obes Relat Metab Disord. 2002. PMID: 11791142 - Mitochondrial DNA content in human omental adipose tissue.
Lindinger A, Peterli R, Peters T, Kern B, von Flüe M, Calame M, Hoch M, Eberle AN, Lindinger PW. Lindinger A, et al. Obes Surg. 2010 Jan;20(1):84-92. doi: 10.1007/s11695-009-9987-3. Epub 2009 Oct 14. Obes Surg. 2010. PMID: 19826890 - Regional variation in adipose tissue insulin action and GLUT4 glucose transporter expression in severely obese premenopausal women.
Marette A, Mauriège P, Marcotte B, Atgié C, Bouchard C, Thériault G, Bukowiecki LJ, Marceau P, Biron S, Nadeau A, Després JP. Marette A, et al. Diabetologia. 1997 May;40(5):590-8. doi: 10.1007/s001250050720. Diabetologia. 1997. PMID: 9165229 - What distinguishes adipose tissue of severely obese humans who are insulin sensitive and resistant?
Xu XJ, Pories WJ, Dohm LG, Ruderman NB. Xu XJ, et al. Curr Opin Lipidol. 2013 Feb;24(1):49-56. doi: 10.1097/MOL.0b013e32835b465b. Curr Opin Lipidol. 2013. PMID: 23298959 Free PMC article. Review.
Cited by
- Adipose Tissue Dysfunction in Obesity: Role of Mineralocorticoid Receptor.
Parasiliti-Caprino M, Bollati M, Merlo FD, Ghigo E, Maccario M, Bo S. Parasiliti-Caprino M, et al. Nutrients. 2022 Nov 9;14(22):4735. doi: 10.3390/nu14224735. Nutrients. 2022. PMID: 36432422 Free PMC article. Review. - Obesity-related insulin resistance: implications for the surgical patient.
Tewari N, Awad S, Macdonald IA, Lobo DN. Tewari N, et al. Int J Obes (Lond). 2015 Nov;39(11):1575-88. doi: 10.1038/ijo.2015.100. Epub 2015 Jun 1. Int J Obes (Lond). 2015. PMID: 26028059 Review. - Malic Enzyme 1 Absence in Synovial Sarcoma Shifts Antioxidant System Dependence and Increases Sensitivity to Ferroptosis Induction with ACXT-3102.
Brashears CB, Prudner BC, Rathore R, Caldwell KE, Dehner CA, Buchanan JL, Lange SES, Poulin N, Sehn JK, Roszik J, Spitzer D, Jones KB, O'Keefe R, Nielsen TO, Taylor EB, Held JM, Hawkins W, Van Tine BA. Brashears CB, et al. Clin Cancer Res. 2022 Aug 15;28(16):3573-3589. doi: 10.1158/1078-0432.CCR-22-0470. Clin Cancer Res. 2022. PMID: 35421237 Free PMC article. - Assessing Obesity-Related Adipose Tissue Disease (OrAD) to Improve Precision Medicine for Patients Living With Obesity.
Pincu Y, Yoel U, Haim Y, Makarenkov N, Maixner N, Shaco-Levy R, Bashan N, Dicker D, Rudich A. Pincu Y, et al. Front Endocrinol (Lausanne). 2022 Apr 29;13:860799. doi: 10.3389/fendo.2022.860799. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35574032 Free PMC article. Review. - The macrophage at the intersection of immunity and metabolism in obesity.
Samaan MC. Samaan MC. Diabetol Metab Syndr. 2011 Oct 28;3(1):29. doi: 10.1186/1758-5996-3-29. Diabetol Metab Syndr. 2011. PMID: 22035457 Free PMC article.
References
- Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence and trends in obesity among US adults, 1999-2000. Jama. 2002;288:1723–1727. - PubMed
- Liang CP, Han S, Senokuchi T, Tall AR. The macrophage at the crossroads of insulin resistance and atherosclerosis. Circ Res. 2007;100:1546–1555. - PubMed
- Reaven G. All obese individuals are not created equal: insulin resistance is the major determinant of cardiovascular disease in overweight/obese individuals. Diab Vasc Dis Res. 2005;2:105–112. - PubMed
- Sims EA. Are there persons who are obese, but metabolically healthy? Metabolism. 2001;50:1499–1504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK032520/DK/NIDDK NIH HHS/United States
- DK32520/DK/NIDDK NIH HHS/United States
- R37 DK030898/DK/NIDDK NIH HHS/United States
- DK30898/DK/NIDDK NIH HHS/United States
- R01 DK030898/DK/NIDDK NIH HHS/United States
- KL2 RR031981/RR/NCRR NIH HHS/United States
- UL1 RR031982/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases