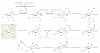Active DNA demethylation: many roads lead to Rome - PubMed (original) (raw)
Review
Active DNA demethylation: many roads lead to Rome
Susan C Wu et al. Nat Rev Mol Cell Biol. 2010 Sep.
Erratum in
- Nat Rev Mol Cell Biol. 2010 Oct;11(10):750
Abstract
DNA methylation is one of the best-characterized epigenetic modifications and has been implicated in numerous biological processes, including transposable element silencing, genomic imprinting and X chromosome inactivation. Compared with other epigenetic modifications, DNA methylation is thought to be relatively stable. Despite its role in long-term silencing, DNA methylation is more dynamic than originally thought as active DNA demethylation has been observed during specific stages of development. In the past decade, many enzymes have been proposed to carry out active DNA demethylation and growing evidence suggests that, depending on the context, this process may be achieved by multiple mechanisms. Insight into how DNA methylation is dynamically regulated will broaden our understanding of epigenetic regulation and have great implications in somatic cell reprogramming and regenerative medicine.
Figures
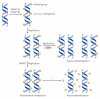
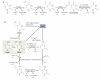
Figure 1. Mechanisms of DNa methylation and demethylation
During early development, methylation patterns are initially established by the de novo DNA methyltransferases DNMT3A and DNMT3B. When DNA replication and cell division occur, these methyl marks are maintained in daughter cells by the maintenance methyltransferase, DNMT1, which has a preference for hemi-methylated DNA. If DNMT1 is inhibited or absent when the cell divides, the newly synthesized strand of DNA will not be methylated and successive rounds of cell division will result in passive demethylation. By contrast, active demethylation can occur through the enzymatic replacement of 5-methylcytosine (5meC) with C.
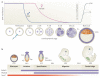
Figure 2. Dynamics of DNa methylation during development
a | Active demethylation in the zygotic paternal genome. Shortly after a sperm fertilizes an egg, the paternal genome rapidly undergoes genome-wide active DNA demethylation and remains demethylated following multiple rounds of cell division. During this time, the maternal genome experiences gradual, passive demethylation. De novo methylation patterns are established by the DNA methyltransferases DNMT3A and DNMT3B during the development of the blastocyst. b | Active demethylation in primordial germ cells (PGCs). After implantation of the blastocyst at embryonic day 7.5 (E7.5), the extraembryonic ectoderm (ExE) and visceral endoderm (VE) produce signals that specify a subset of epiblast cells (Epi) to become PGCs. This process requires two key transcription factors, BLIMP1 (also known as PR domain zinc finger protein 1 (PRDM1)) and PDRM14, which are expressed during this stage of development. Following specification, PGC founder cells divide in the presence of the DNA methyltransferase DNMT1 and migrate towards the genital ridge. During this migration and on arrival at the genital ridge, 5-methylcytosine (5meC) is erased through an active mechanism. ICM, inner cell mass; TE, trophectoderm.
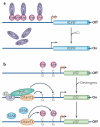
Figure 3. Locus-specific active DNa demethylation in somatic cells
a | Active demethylation at the brain-derived neurotrophic factor (BDNF) promoter. In neurons, BDNF is maintained in a repressed state through DNA methylation and binding of the repressive methylcytosine (meC)-binding protein MeCP2. On depolarization with KCl, DNA methylation and MeCP2 binding are lost, concomitant with increased BDNF expression. This demethylation event is considered to be active because it occurs in post-mitotic neurons. b | Active demethylation at nuclear receptor target promoters. The promoter of the oestrogen receptor (ER) target gene pS2 (also known as TFF1) undergoes cyclical rounds of methylation and demethylation that correspond to the repression and expression of the gene, respectively. Transcriptional activation of pS2 occurs in the presence of oestrogens (E2) and coincides with demethylation of the promoter. This is achieved by deamination of 5meC by DNA methyltransferase 3 (DNMT3) followed by base excision repair (BER) of the T•G mismatch by T DNA glycosylase (TDG). To revert to repression, DNMT3 re-methylates the promoter. Although DNMT3 is involved in both methylation and demethylation, it is important to note that DNMT3 can only carry out the deamination step in the absence or at low concentrations of the methyl donor _S_-adenosylmethionine (SAM).
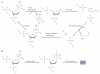
Figure 4. Base excision repair-based mechanisms for DNa demethylation
a | Base excision repair (BER) through direct excision of 5-methylcytosine (5meC). Initiation of the BER pathway can be carried out by a glycosylase that directly excises 5meC to generate an abasic (apurinic and apyrimidinic (AP)) site. The DNA backbone is nicked by an AP lyase (or by the glycosylase itself if it is bifunctional). The 3′ sugar group is then cleaved by an AP endonuclease and the resulting single nucleotide gap is filled in with an unmethylated C by an unknown polymerase and ligase. It has been well established in plants that the demeter (Dme; also known as repressor of silencing 1 (ROS1)) family of enzymes can carry out the 5meC glycosylase reaction, but to date no mammalian enzymes have been reported to be capable of carrying out this step efficiently. b | Deamination of 5meC followed by BER. In contrast to direct excision of 5meC, deamination of 5meC produces T, which can be repaired by BER by a T•G mismatch glycosylase such as T DNA glycosylase (TDG) or methyl-CpG-binding domain protein 4 (MBD4) to regenerate an unmethylated C. DNMT, DNA methyltransferase.
Figure 5. Oxidative demethylation by TET proteins
a | Part of the thymidine salvage pathway. Direct removal of the methyl group of 5-methylcytosine (5meC) involves breaking a carbon-carbon bond, which requires an enzyme with great catalytic power. Such an enzyme exists in the thymidine salvage pathway. Starting with T, thymine-7-hydroxylase (THase) carries out three consecutive hydroxylation reactions to produce iso-orotate, which is processed by a decarboxylase to produce U. A similar mechanism may be used in active DNA demethylation, particularly by the ten-eleven translocation (TET) family of proteins. b | The fate of 5-hydroxymethylcytosine (5hmC). The TET family of proteins catalyses the conversion of 5meC to 5hmC, which may be an intermediate that can be further processed by one of the following mechanisms. BER may be initiated by a 5hmC glycosylase (1); 5hmC may undergo deamination to produce 5hmU (2), which is repaired by BER through a 5hmU glycosylase such as SMUG1 (single-strand-selective monofunctional U DNA glycosylase 1); 5hmC may directly be converted to C by DNA methyltransferases (DNMTs), ultraviolet (UV) exposure or high pH (3); or consecutive hydroxylation reactions followed by a decarboxylation reaction similar to the thymidine salvage pathway may be used to ultimately replace 5hmC with C (4). Alternatively, 5hmC itself may be a functional modification. α-KG, α-ketoglutarate.
Figure 6. Proposed mechanism for elP3-mediated DNa demethylation
Mammalian elongator complex protein 3 (ELP3) contains an Fe-S radical S-adenosylmethionine (SAM) domain that is important for active DNA demethylation of the zygotic paternal genome. If ELP3 is indeed a functional radical SAM protein, it may directly carry out DNA demethylation through the following mechanism. First, ELP3 uses SAM to generate a 5′-deoxyadenosyl radical, which could extract a hydrogen atom from the 5-methyl group of 5-methylcytosine (5meC; 1) to form a 5meC radical (2). After an electron is donated back to the Fe -S to create the third intermediate (3), a water molecule would promote the formation of 5-hydroxymethylcytosine (5hmC) (4). A nucleophilic attack at carbon 6 can result in the carbon-carbon bond breaking to release formaldehyde (5-7). In the absence of an external nucleophile, an alternative pathway (4′-6′) that leads to the release of formaldehyde can also take place. Finally, an elimination step would produce an end product of C (8).
Similar articles
- [Role of 5-hydroxymethylcytosine and TET proteins in epigenetic regulation of gene expression].
Głowacki S, Błasiak J. Głowacki S, et al. Postepy Biochem. 2013;59(1):64-9. Postepy Biochem. 2013. PMID: 23821944 Review. Polish. - Active DNA demethylation by DNA repair: Facts and uncertainties.
Schuermann D, Weber AR, Schär P. Schuermann D, et al. DNA Repair (Amst). 2016 Aug;44:92-102. doi: 10.1016/j.dnarep.2016.05.013. Epub 2016 May 16. DNA Repair (Amst). 2016. PMID: 27247237 Review. - [Roles of ten eleven translocation proteins family and 5-hydroxymethylcytosine in epigenetic regulation of stem cells and regenerative medicine].
Zhao JF, Li D, An Y. Zhao JF, et al. Beijing Da Xue Xue Bao Yi Xue Ban. 2021 Feb 22;53(2):420-424. doi: 10.19723/j.issn.1671-167X.2021.02.032. Beijing Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 33879920 Free PMC article. Chinese. - MicroRNAs mediated targeting on the Yin-yang dynamics of DNA methylation in disease and development.
Tu J, Liao J, Luk AC, Tang NL, Chan WY, Lee TL. Tu J, et al. Int J Biochem Cell Biol. 2015 Oct;67:115-20. doi: 10.1016/j.biocel.2015.05.002. Epub 2015 May 12. Int J Biochem Cell Biol. 2015. PMID: 25979370 Review. - Epigenetic reprogramming in mammalian development.
Reik W, Dean W, Walter J. Reik W, et al. Science. 2001 Aug 10;293(5532):1089-93. doi: 10.1126/science.1063443. Science. 2001. PMID: 11498579 Review.
Cited by
- 5-Hydroxymethylcytosine: Far Beyond the Intermediate of DNA Demethylation.
Zheng K, Lyu Z, Chen J, Chen G. Zheng K, et al. Int J Mol Sci. 2024 Nov 2;25(21):11780. doi: 10.3390/ijms252111780. Int J Mol Sci. 2024. PMID: 39519332 Free PMC article. Review. - Methylation of T and B Lymphocytes in Autoimmune Rheumatic Diseases.
Deng T, Wang Z, Geng Q, Wang Z, Jiao Y, Diao W, Xu J, Deng T, Luo J, Tao Q, Xiao C. Deng T, et al. Clin Rev Allergy Immunol. 2024 Jun;66(3):401-422. doi: 10.1007/s12016-024-09003-4. Epub 2024 Aug 29. Clin Rev Allergy Immunol. 2024. PMID: 39207646 Review. - Whole-Genome Bisulfite Sequencing Protocol for the Analysis of Genome-Wide DNA Methylation and Hydroxymethylation Patterns at Single-Nucleotide Resolution.
Derbala D, Garnier A, Bonnet E, Deleuze JF, Tost J. Derbala D, et al. Methods Mol Biol. 2024;2842:353-382. doi: 10.1007/978-1-0716-4051-7_18. Methods Mol Biol. 2024. PMID: 39012605 - DNA methylation modification in Idiopathic pulmonary fibrosis.
Ren L, Chang YF, Jiang SH, Li XH, Cheng HP. Ren L, et al. Front Cell Dev Biol. 2024 Jun 10;12:1416325. doi: 10.3389/fcell.2024.1416325. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38915445 Free PMC article. Review. - The pharmacoepigenetic paradigm in cancer treatment.
Ocaña-Paredes B, Rivera-Orellana S, Ramírez-Sánchez D, Montalvo-Guerrero J, Freire MP, Espinoza-Ferrao S, Altamirano-Colina A, Echeverría-Espinoza P, Ramos-Medina MJ, Echeverría-Garcés G, Granda-Moncayo D, Jácome-Alvarado A, Andrade MG, López-Cortés A. Ocaña-Paredes B, et al. Front Pharmacol. 2024 Apr 24;15:1381168. doi: 10.3389/fphar.2024.1381168. eCollection 2024. Front Pharmacol. 2024. PMID: 38720770 Free PMC article. Review.
References
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 2003;33:245–254. - PubMed
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nature Rev. Genet. 2007;8:286–298. - PubMed
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Rev. Cancer. 2004;4:143–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous