Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex - PubMed (original) (raw)
Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex
F Javier Pérez-Victoria et al. Mol Biol Cell. 2010.
Abstract
The Golgi-associated retrograde protein (GARP) complex mediates tethering and fusion of endosome-derived transport carriers to the trans-Golgi network (TGN). In the yeast Saccharomyces cerevisiae, GARP comprises four subunits named Vps51p, Vps52p, Vps53p, and Vps54p. Orthologues of the GARP subunits, except for Vps51p, have been identified in all other eukaryotes. A yeast two-hybrid screen of a human cDNA library yielded a phylogenetically conserved protein, Ang2/Fat-free, which interacts with human Vps52, Vps53 and Vps54. Human Ang2 is larger than yeast Vps51p, but exhibits significant homology in an N-terminal coiled-coil region that mediates assembly with other GARP subunits. Biochemical analyses show that human Ang2, Vps52, Vps53 and Vps54 form an obligatory 1:1:1:1 complex that strongly interacts with the regulatory Habc domain of the TGN SNARE, Syntaxin 6. Depletion of Ang2 or the GARP subunits similarly impairs protein retrieval to the TGN, lysosomal enzyme sorting, endosomal cholesterol traffic¤ and autophagy. These findings indicate that Ang2 is the missing component of the GARP complex in most eukaryotes.
Figures
Figure 1.
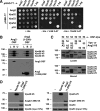
Alignment and coiled-coil prediction of Ang2 orthologues. (A) Amino acid sequence alignment of the N-terminal regions of human (Homo sapiens, Hs; AAF21627), zebrafish (Danio rerio, Dr; NP_001036200), Drosophila melanogaster (Dm; NP_611363), Caenorhabditis elegans (Ce; NP_001020972) and Arabidopsis thaliana (At; NP_192112) Ang2, and of full-length Yarrowia lipolytica (Yl; XP_500348) and S. cerevisiae (Sc; NP_012945) Vps51p. Alignments were performed with CLUSTALW and BOXSHADE using a 0.3 fraction for shading. Black and gray boxes indicate identical and conserved residues, respectively. A longer, 812-amino acid form of human Ang2 (EAW74347) was also found in protein databases. (B) Coiled-coil prediction (Lupas et al., 1991) in full-length human and zebrafish Ang2, and S. cerevisiae Vps51p. The graph displays the probability of coiled-coils (_y_-axis) versus amino acid number (_x_-axis).
Figure 2.
Identification of Ang2 as a component of the human GARP complex. (A) Y2H analysis of binary interactions among human Ang2, Vps52, Vps53, and Vps54. All constructs were full-length, except for Vps54, which comprised residues 1–535 from the 977-residue protein. Two concentrations of 3-AT were used to suppress self-activation by some constructs. The SV40 large T antigen (T Ag) and p53 were used as controls. Growth in the absence of histidine (−His) is indicative of interactions. (B and C) Coassembly of Ang2 with Vps52, Vps53, and Vps54. (B) HeLa cells were cotransfected with plasmids encoding One-STrEP/FLAG (OSF)-tagged human Ang2 _(_full-length or an N-terminal fragment spanning residues 1–299) and V5-tagged versions of Vps52, Vps53, and Vps54. Tags were at the C-termini. Cell extracts were incubated with StrepTactin-Sepharose, and the lysate (L; 5% of the total), bound (B), and unbound (U) fractions were analyzed by SDS-PAGE and immunoblotting with antibodies to the V5 and FLAG tags. (C) HeLa cells were cotransfected with plasmids encoding the combinations of OSF- or V5-tagged Ang2, Vps52, Vps53, and/or Vps54 indicated in the figure. After isolation on StrepTactin-Sepharose, lysate (10% of the total) and bound fractions were analyzed as in C. Molecular-mass markers (in kDa) are indicated on the left. (D) HeLa cells were transfected with an empty vector (−) or plasmids encoding V5-tagged Vps45 (negative control) or Ang2 (1–299). At 12 h after transfection, cells were lysed and subjected to immunoprecipitation with an antibody to the V5 epitope (top panel). Total levels (bottom) and coprecipitation of endogenous Vps52 and Vps53 with Ang2 (1–299; middle) were detected by immunoblotting with antibodies to Vps52 and Vps53, respectively.
Figure 3.
Properties of the complete GARP complex including Ang2. (A) Subcellular fractionation of HeLa cells expressing V5-tagged versions of Ang2, Vps52, Vps53, and Vps54. Postnuclear supernatants were separated by ultracentrifugation into cytosol and membrane fractions. Membranes were extracted with either 1 M NaCl or 1% Triton X-100 and further separated into supernatant (S) and pellet (P) fractions. (B) The supernatant of the NaCl extraction was loaded on a linear 5–20% glycerol gradient and ultracentrifuged for 16 h. Arrows indicate the migration of standard proteins on the gradient (s20,w values in Svedberg units). (C) The same salt extract was analyzed by gel filtration on a Superose 6 column. Arrows indicate the elution positions of molecular size markers (Stokes radii in Å). (A–C) Fractions were analyzed by SDS-PAGE and immunoblotting with antibodies to the V5 epitope, endogenous Vps53, and various proteins used as controls for membrane association (A) or as standards of known hydrodynamic properties (B and C). (D and E) Detergent extracts of HeLa cells expressing V5-tagged versions of Ang2 alone (D), Vps52, Vps53, and Vps54 (E, bottom) or the four proteins together (E, top) were incubated with the indicated GST-SNARE fusion proteins. Bound proteins were analyzed by SDS-PAGE and immunoblotting with antibody to the V5 epitope. In all electrophoretograms, molecular-mass markers (in kDa) are indicated on the left.
Figure 4.
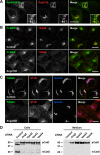
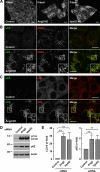
TGN localization and requirement of Ang2 for retrograde transport and acid hydrolase sorting to lysosomes. (A) Colocalization of V5-tagged Ang2 (red) and GFP-tagged Vps54 (green) in HeLa cells stained with antibody to the V5 epitope. (B) Redistribution of endogenous CI-MPR (green) and TGN46 (red) upon Ang2 knockdown (KD) in HeLa cells. Yellow in the merged panels indicates overlapping localization in the green and red channels. (C) Control and Ang2 KD HeLa cells were placed on ice and incubated with 1 μg/ml Cy3-conjugated B-subunit of Shiga toxin (STxB; red) for 30 min and chased in complete medium for 60 min before fixation and immunostaining for TGN46 (green) and giantin (blue). Overlaps in the merged panels are shown in yellow (green and red channels), cyan (green and blue channels), magenta (red and blue channels) and white (all three channels). Imaging was performed by confocal fluorescence microscopy. Bars, 10 μm. (D) Analysis of cathepsin D processing (cells) and secretion (medium) in control HeLa cells or cells depleted of Ang2 or the other GARP subunits. pCatD and mCatD represent precursor and mature cathepsin D, respectively. Molecular-mass markers (in kDa) are indicated on the left.
Figure 5.
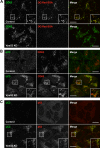
Lysosomal defects in Ang2-KD cells. (A) Endogenous cathepsin D (CatD; green) and CD63 (red) in control and Ang2-KD HeLa cells. (B) Control or Ang2-KD HeLa cells were incubated overnight with 10 μg/ml DQ Red BSA (red) in complete medium before fixation and staining with antibody to CD63 (green). Colocalization is indicated as described in the legend to Figure 4. Imaging was performed by confocal fluorescence microscopy. Bars, 10 μm.
Figure 6.
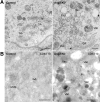
Electron microscopy showing aberrant vacuolar structures in Ang2-KD cells. (A) Ultrathin sections of Epon-embedded control and Ang2-KD cells. (B) Ultrathin frozen sections of control and Ang2-KD cells immunolabeled with antibody to CD63 and protein A-gold (10-nm diameter). GA, Golgi apparatus; End, endosome; m, mitochondria; MVB, multivesicular body; Vac, vacuole. Bars, 500 nm.
Figure 7.
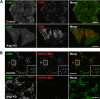
Accumulation of cholesterol and autophagic structures in cells depleted of Ang2 or other GARP subunits. (A) Filipin staining of unesterified cholesterol in control, Ang2-, or Vps52-KD HeLa cells. (B and C) Immunofluorescence microscopy of control or Ang2-KD HeLa cells grown in complete medium and costained with antibodies to LC3 (green) and CD63 (red; B) and LC3 (green) and p62 (red; C). Colocalization is indicated as described in the legend to Figure 4. Bars, 10 μm. (D) Lysates of control or Ang2- or Vps52-KD HeLa cells were analyzed by SDS-PAGE and immunoblotting with antibodies to LC3, p62 and actin. Molecular-mass markers (in kDa) are indicated on the left. (E) Quantification from several experiments such as that in D. Values are the mean ± SD from four independent experiments. **p < 0.01.
Figure 8.
Lysosomal and autophagic dysfunction in Vps52-KD cells. (A–C) Immunofluorescence microscopy of control or Vps52-KD HeLa cells grown in complete medium and costained with antibodies to CD63 (green; A), LC3 (green) and CD63 (red; B), and LC3 (green) and p62 (red; C). In A, cells were incubated overnight with 10 μg/ml DQ Red BSA (red) in complete medium before fixation and immunostaining. Colocalization is indicated as described in the legend to Figure 4. Bars, 10 μm.
Similar articles
- The Caenorhabditis elegans GARP complex contains the conserved Vps51 subunit and is required to maintain lysosomal morphology.
Luo L, Hannemann M, Koenig S, Hegermann J, Ailion M, Cho MK, Sasidharan N, Zweckstetter M, Rensing SA, Eimer S. Luo L, et al. Mol Biol Cell. 2011 Jul 15;22(14):2564-78. doi: 10.1091/mbc.E10-06-0493. Epub 2011 May 25. Mol Biol Cell. 2011. PMID: 21613545 Free PMC article. - TSSC1 is novel component of the endosomal retrieval machinery.
Gershlick DC, Schindler C, Chen Y, Bonifacino JS. Gershlick DC, et al. Mol Biol Cell. 2016 Sep 15;27(18):2867-78. doi: 10.1091/mbc.E16-04-0209. Epub 2016 Jul 20. Mol Biol Cell. 2016. PMID: 27440922 Free PMC article. - Structural basis for the interaction of the Golgi-Associated Retrograde Protein Complex with the t-SNARE Syntaxin 6.
Abascal-Palacios G, Schindler C, Rojas AL, Bonifacino JS, Hierro A. Abascal-Palacios G, et al. Structure. 2013 Sep 3;21(9):1698-706. doi: 10.1016/j.str.2013.06.025. Epub 2013 Aug 8. Structure. 2013. PMID: 23932592 Free PMC article. - Transport according to GARP: receiving retrograde cargo at the trans-Golgi network.
Bonifacino JS, Hierro A. Bonifacino JS, et al. Trends Cell Biol. 2011 Mar;21(3):159-67. doi: 10.1016/j.tcb.2010.11.003. Epub 2010 Dec 21. Trends Cell Biol. 2011. PMID: 21183348 Free PMC article. Review. - Role of GARP Vesicle Tethering Complex in Golgi Physiology.
Khakurel A, Lupashin VV. Khakurel A, et al. Int J Mol Sci. 2023 Mar 23;24(7):6069. doi: 10.3390/ijms24076069. Int J Mol Sci. 2023. PMID: 37047041 Free PMC article. Review.
Cited by
- Identification of shared genetic architecture between non-alcoholic fatty liver disease and type 2 diabetes: A genome-wide analysis.
Tan Y, He Q, Chan KHK. Tan Y, et al. Front Endocrinol (Lausanne). 2023 Mar 22;14:1050049. doi: 10.3389/fendo.2023.1050049. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37033223 Free PMC article. - Zebrafish: an important tool for liver disease research.
Goessling W, Sadler KC. Goessling W, et al. Gastroenterology. 2015 Nov;149(6):1361-77. doi: 10.1053/j.gastro.2015.08.034. Epub 2015 Aug 28. Gastroenterology. 2015. PMID: 26319012 Free PMC article. Review. - A neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes.
Gershlick DC, Ishida M, Jones JR, Bellomo A, Bonifacino JS, Everman DB. Gershlick DC, et al. Hum Mol Genet. 2019 May 1;28(9):1548-1560. doi: 10.1093/hmg/ddy423. Hum Mol Genet. 2019. PMID: 30624672 Free PMC article. - Autophagic substrate clearance requires activity of the syntaxin-5 SNARE complex.
Renna M, Schaffner C, Winslow AR, Menzies FM, Peden AA, Floto RA, Rubinsztein DC. Renna M, et al. J Cell Sci. 2011 Feb 1;124(Pt 3):469-82. doi: 10.1242/jcs.076489. J Cell Sci. 2011. PMID: 21242315 Free PMC article. - MTV proteins unveil ER- and microtubule-associated compartments in the plant vacuolar trafficking pathway.
Delgadillo MO, Ruano G, Zouhar J, Sauer M, Shen J, Lazarova A, Sanmartín M, Lai LTF, Deng C, Wang P, Hussey PJ, Sánchez-Serrano JJ, Jiang L, Rojo E. Delgadillo MO, et al. Proc Natl Acad Sci U S A. 2020 May 5;117(18):9884-9895. doi: 10.1073/pnas.1919820117. Epub 2020 Apr 22. Proc Natl Acad Sci U S A. 2020. PMID: 32321832 Free PMC article.
References
- Ballabio A. Disease pathogenesis explained by basic science: lysosomal storage diseases as autophagocytic disorders. Int. J. Clin. Pharmacol. Ther. 2009;47(Suppl 1):S34–S38. - PubMed
- Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous