SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome - PubMed (original) (raw)
. 2010 Oct 15;10(8):796-810.
doi: 10.4161/cbt.10.8.12914. Epub 2010 Oct 15.
H C Harsha, Santosh Renuse, Harsh Pawar, Nandini A Sahasrabuddhe, Min-Sik Kim, Arivusudar Marimuthu, Shivakumar Keerthikumar, Babylakshmi Muthusamy, Kumaran Kandasamy, Yashwanth Subbannayya, Thottethodi Subrahmanya Keshava Prasad, Riaz Mahmood, Raghothama Chaerkady, Stephen J Meltzer, Rekha V Kumar, Anil K Rustgi, Akhilesh Pandey
Affiliations
- PMID: 20686364
- PMCID: PMC3093916
- DOI: 10.4161/cbt.10.8.12914
SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome
Manoj Kumar Kashyap et al. Cancer Biol Ther. 2010.
Abstract
The identification of secreted proteins that are differentially expressed between non-neoplastic and esophageal squamous cell carcinoma (ESCC) cells can provide potential biomarkers of ESCC. We used a SILAC-based quantitative proteomic approach to compare the secretome of ESCC cells with that of non-neoplastic esophageal squamous epithelial cells. Proteins were resolved by SDS-PAGE, and tandem mass spectrometry analysis (LC-MS/MS) of in-gel trypsin-digested peptides was carried out on a high-accuracy qTOF mass spectrometer. In total, we identified 441 proteins in the combined secretomes, including 120 proteins with > 2-fold upregulation in the ESCC secretome vs. that of non-neoplastic esophageal squamous epithelial cells. In this study, several potential protein biomarkers previously known to be increased in ESCC including matrix metalloproteinase 1, transferrin receptor, and transforming growth factor beta-induced 68 kDa were identified as overexpressed in the ESCC-derived secretome. In addition, we identified several novel proteins that have not been previously reported to be associated with ESCC. Among the novel candidate proteins identified, protein disulfide isomerase family a member 3 (PDIA3), GDP dissociation inhibitor 2 (GDI2), and lectin galactoside binding soluble 3 binding protein (LGALS3BP) were further validated by immunoblot analysis and immunohistochemical labeling using tissue microarrays. This tissue microarray analysis showed overexpression of protein disulfide isomerase family a member 3, GDP dissociation inhibitor 2, and lectin galactoside binding soluble 3 binding protein in 93%, 93% and 87% of 137 ESCC cases, respectively. Hence, we conclude that these potential biomarkers are excellent candidates for further evaluation to test their role and efficacy in the early detection of ESCC.
Figures
Figure 1
The work flow for discovery and initial validation of biomarkers for esophageal squamous cell carcinoma. For SILAC labeling, Het-1A cells were grown in ‘heavy’ medium and the TE-series ESCC cells were grown in ‘light’ medium as indicated. The secretome was normalized, pooled and resolved by SDS-PAGE. Gel bands were excised and in-gel trypsin digested followed by liquid chromatography tandem mass spectrometry (LC-MS/MS) on a qTOF mass spectrometer. The data was searched using Mascot and Spectrum Mill search engines. Some of the overexpressed proteins that were not previously described (e.g., PDIA3) were validated using western blot and IHC labeling using tissue microarrays.
Figure 2
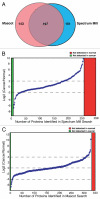
Protein profiled using the SILAC strategy. (A) Venn diagram showing proteins identified by Mascot and Spectrum Mill search algorithms. (B) Distribution of proteins identified by Spectrum Mill plotted against the log2 ratios as indicated. Proteins for which peptides were only observed in cancer cell lines are indicated in red while those for which peptides were identified only in the normal cell line are shown in green. (C) Distribution of proteins identified by Mascot plotted against the log2 ratios as indicated. Proteins for which peptides were only observed in cancer cell lines are indicated in red while those for which peptides were identified only in the normal cell line are shown in green.
Figure 3
MS and MS/MS spectra of selected differentially expressed proteins. MS and MS/MS spectra of peptide from representative differentially expressed proteins identified in this study. (A) Matrix metalloproteinase 1 (MMP1); (B) TGFbeta induced, 68 KD (TGFBI); (C) GDP dissociation inhibitor 2 (GDI2); (D) Protein disulfide isomerase A3 (PDIA3); (E) Nicotinamide phosphoribosyltransferase (NAMPT) and (F) Lectin, galactoside-binding, soluble, 3 binding protein (LGALS3BP).
Figure 4
Western blot validation for selected proteins identified in the ESCC secretome. Pooled conditioned media from different ESCC cell lines and normal cell line was tested for expression of the indicated proteins using commercially available antibodies.
Figure 5
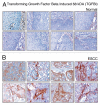
Validation of TGFBI using immunohistochemical labeling. Expression of TGFBI in representative normal esophageal squamous mucosa (A). Expression of TGFBI in ESCC is observed in both stromal and epithelial cell compartments (B).
Figure 6
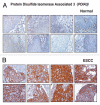
Validation of PDIA3 using immunohistochemical labeling. Expression of PDIA3 in representative normal esophageal squamous mucosa (A). Expression of PDIA3 in ESCC is observed in both stromal and epithelial cell compartments (B).
Figure 7
Validation of LGALS3BP using immunohistochemical labeling. Expression of LGALS3BP in representative normal esophageal squamous mucosa (A). Expression of LGALS3BP in ESCC is observed in both stromal and epithelial compartments (B).
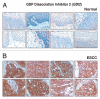
Figure 8
Validation of GDI2 using immunohistochemical labeling. Expression of GDI2 in representative normal esophageal squamous mucosa (A). Expression of GDI2 in ESCC is observed in both stromal and epithelial compartments (B).
Comment in
- Maximizing early detection of esophageal squamous cell carcinoma via SILAC-proteomics.
Lee KK, Todorova K, Mandinova A. Lee KK, et al. Cancer Biol Ther. 2010 Oct 15;10(8):811-3. doi: 10.4161/cbt.10.8.13754. Epub 2010 Oct 15. Cancer Biol Ther. 2010. PMID: 20953140 Free PMC article. No abstract available.
Similar articles
- Maximizing early detection of esophageal squamous cell carcinoma via SILAC-proteomics.
Lee KK, Todorova K, Mandinova A. Lee KK, et al. Cancer Biol Ther. 2010 Oct 15;10(8):811-3. doi: 10.4161/cbt.10.8.13754. Epub 2010 Oct 15. Cancer Biol Ther. 2010. PMID: 20953140 Free PMC article. No abstract available. - Quantitative tissue proteomics of esophageal squamous cell carcinoma for novel biomarker discovery.
Pawar H, Kashyap MK, Sahasrabuddhe NA, Renuse S, Harsha HC, Kumar P, Sharma J, Kandasamy K, Marimuthu A, Nair B, Rajagopalan S, Maharudraiah J, Premalatha CS, Kumar KV, Vijayakumar M, Chaerkady R, Prasad TS, Kumar RV, Kumar RV, Pandey A. Pawar H, et al. Cancer Biol Ther. 2011 Sep 15;12(6):510-22. doi: 10.4161/cbt.12.6.16833. Epub 2011 Sep 15. Cancer Biol Ther. 2011. PMID: 21743296 Free PMC article. - Identification of galectin-7 as a potential biomarker for esophageal squamous cell carcinoma by proteomic analysis.
Zhu X, Ding M, Yu ML, Feng MX, Tan LJ, Zhao FK. Zhu X, et al. BMC Cancer. 2010 Jun 15;10:290. doi: 10.1186/1471-2407-10-290. BMC Cancer. 2010. PMID: 20546628 Free PMC article. - An overview of esophageal squamous cell carcinoma proteomics.
Qi YJ, Chao WX, Chiu JF. Qi YJ, et al. J Proteomics. 2012 Jun 18;75(11):3129-37. doi: 10.1016/j.jprot.2012.04.025. Epub 2012 Apr 27. J Proteomics. 2012. PMID: 22564818 Review. - Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes.
Munday DC, Surtees R, Emmott E, Dove BK, Digard P, Barr JN, Whitehouse A, Matthews D, Hiscox JA. Munday DC, et al. Proteomics. 2012 Feb;12(4-5):666-72. doi: 10.1002/pmic.201100488. Epub 2012 Jan 19. Proteomics. 2012. PMID: 22246955 Review.
Cited by
- Methodologies to decipher the cell secretome.
Mukherjee P, Mani S. Mukherjee P, et al. Biochim Biophys Acta. 2013 Nov;1834(11):2226-32. doi: 10.1016/j.bbapap.2013.01.022. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376189 Free PMC article. Review. - Expression, regulation and targeting of receptor tyrosine kinases in esophageal squamous cell carcinoma.
Kashyap MK, Abdel-Rahman O. Kashyap MK, et al. Mol Cancer. 2018 Feb 19;17(1):54. doi: 10.1186/s12943-018-0790-4. Mol Cancer. 2018. PMID: 29455652 Free PMC article. Review. - Mass spectrometry-based proteomics in molecular diagnostics: discovery of cancer biomarkers using tissue culture.
Paul D, Kumar A, Gajbhiye A, Santra MK, Srikanth R. Paul D, et al. Biomed Res Int. 2013;2013:783131. doi: 10.1155/2013/783131. Epub 2013 Mar 17. Biomed Res Int. 2013. PMID: 23586059 Free PMC article. Review. - SILAC-based quantitative MS approach for real-time recording protein-mediated cell-cell interactions.
Wang X, He Y, Ye Y, Zhao X, Deng S, He G, Zhu H, Xu N, Liang S. Wang X, et al. Sci Rep. 2018 May 31;8(1):8441. doi: 10.1038/s41598-018-26262-2. Sci Rep. 2018. PMID: 29855483 Free PMC article. - A Sensitized Screen for Genes Promoting Invadopodia Function In Vivo: CDC-42 and Rab GDI-1 Direct Distinct Aspects of Invadopodia Formation.
Lohmer LL, Clay MR, Naegeli KM, Chi Q, Ziel JW, Hagedorn EJ, Park JE, Jayadev R, Sherwood DR. Lohmer LL, et al. PLoS Genet. 2016 Jan 14;12(1):e1005786. doi: 10.1371/journal.pgen.1005786. eCollection 2016 Jan. PLoS Genet. 2016. PMID: 26765257 Free PMC article.
References
- Oka D, Yamashita S, Tomioka T, Nakanishi Y, Kato H, Kaminishi M, Ushijima T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115:3412–3426. - PubMed
- Yu C, Chen K, Zheng H, Guo X, Jia W, Li M, Zeng M, Li J, Song L. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK087454/DK/NIDDK NIH HHS/United States
- R01 DK087454/DK/NIDDK NIH HHS/United States
- CA85069/CA/NCI NIH HHS/United States
- CA146799/CA/NCI NIH HHS/United States
- P01-CA098101/CA/NCI NIH HHS/United States
- P01 CA098101/CA/NCI NIH HHS/United States
- U01 CA085069/CA/NCI NIH HHS/United States
- R01 CA146799/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous