Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria - PubMed (original) (raw)
Interaction and modulation of two antagonistic cell wall enzymes of mycobacteria
Erik C Hett et al. PLoS Pathog. 2010.
Abstract
Bacterial cell growth and division require coordinated cell wall hydrolysis and synthesis, allowing for the removal and expansion of cell wall material. Without proper coordination, unchecked hydrolysis can result in cell lysis. How these opposing activities are simultaneously regulated is poorly understood. In Mycobacterium tuberculosis, the resuscitation-promoting factor B (RpfB), a lytic transglycosylase, interacts and synergizes with Rpf-interacting protein A (RipA), an endopeptidase, to hydrolyze peptidoglycan. However, it remains unclear what governs this synergy and how it is coordinated with cell wall synthesis. Here we identify the bifunctional peptidoglycan-synthesizing enzyme, penicillin binding protein 1 (PBP1), as a RipA-interacting protein. PBP1, like RipA, localizes both at the poles and septa of dividing cells. Depletion of the ponA1 gene, encoding PBP1 in M. smegmatis, results in a severe growth defect and abnormally shaped cells, indicating that PBP1 is necessary for viability and cell wall stability. Finally, PBP1 inhibits the synergistic hydrolysis of peptidoglycan by the RipA-RpfB complex in vitro. These data reveal a post-translational mechanism for regulating cell wall hydrolysis and synthesis through protein-protein interactions between enzymes with antagonistic functions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. M. tuberculosis RipA identifies bifunctional synthase PBP1.
(A) RipA, a 472 residue protein, contains a domain of unknown function (COG3883) as well as a predicted endopeptidase domain. The region used to screen for interacting proteins in a yeast two-hybrid screen is shown, consisting of amino acids 350–472. (B) PBP1, a 678 residue protein encoded by ponA1, is a bifunctional peptidoglycan synthase. PBP1 contains a transglycosylase domain at the N-terminus and a penicillin-sensitive transpeptidase domain at the C-terminus.
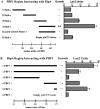
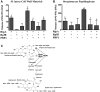
Figure 2. C-terminal regions are required for interaction.
(A) Data from quantitative LacZ assays of four overlapping regions of 200 amino acids each of PBP1 tested for interaction with the C-terminal 123 amino acids of RipA. The C-terminal 259 amino acids of PBP1 were found to be sufficient for interaction with RipA, while regions lacking the C-terminal 150 amino acids failed to interact. Data shown are from a representative experiment done in triplicate. Data are represented as mean +/− SEM. (B) Data from quantitative LacZ assays of several different deletion constructs for interaction with the C-terminal 259 amino acids of PBP1 in the yeast two-hybrid assay. Deletion of the C-terminal of RipA decreased the intensity of interaction and the C-terminal 25 amino acids of RipA were sufficient for interaction with PBP1. Data shown are from a representative experiment done in triplicate. Data are represented as mean +/− SEM.
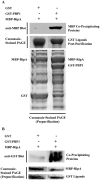
Figure 3. Recombinant PBP1 coprecipitates with RipA in vitro.
(A) Fusion proteins were co-expressed in E. coli and GST fusion proteins were purified directly from the lysate. Co-purifying MBP fusion proteins were detected by Western blotting using anti-MBP antibody (top panel). Unfused GST was used to test the specificity of the interaction. A Coomassie-stained PAGE gel containing lysates obtained prior to GST purification (bottom panel) and after GST purification (middle panel) is shown to demonstrate that similar amounts of proteins were available for pulldown. (B) Proteins were separately purified from E. coli, combined as indicated in equimolar amounts, incubated, then purified on amylose resin. Samples were taken before (middle and bottom panels) and after (top panel) MBP purification. A Coomassie-stained PAGE gel containing protein mixtures prior to MBP purification is shown to demonstrate that similar amounts of proteins were available for pulldown. Co-purifying GST fusion proteins were detected by Western blotting using anti-GST antibody. Unfused GST was used to test the specificity of the interaction.
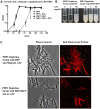
Figure 4. PBP1 localizes to the poles and septa of M. smegmatis.
Fluorescence microscopy of M. smegmatis expressing M. tuberculosis PBP1 fused to monomeric red fluorescent protein (RFP). PBP1-RFP fusion protein is under the control of a tetracycline-inducible promoter. PBP1 localized to the poles and also to septa. Arrows indicate narrow septa where PBP1 does not localize and arrowheads indicate larger septa where PBP1 localizes.
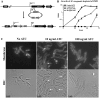
Figure 5. Depletion of PBP1 blocks cell division.
(A) Diagram showing the strategy used to replace the native promoter of the ponA1 operon (PponA1) in M. smegmatis with a tetracycline-inducible promoter (Ptet) through homologous recombination (strategy and diagram adapted from [12]). OriE: E. coli origin of replication. (B) PBP1 (ponA1) depletion strain of M. smegmatis was grown with inducer (ATC = anhydrotetracycline), then inoculated into media with decreasing amounts of inducer and followed by OD600 over time. Data are represented as mean +/− standard deviation. (C) Micrographs of M. smegmatis PBP1 (ponA1) depletion strain with membranes imaged by staining with TMA-DPH. Bacteria were grown with no anhydrotetracycline inducer (No ATC), 10, or 100 ng/ml ATC. Bacteria grown with 100 ng/ml ATC grew as wildtype, while No ATC and 10 ng/ml ATC grew slowly, with bulbous poles and round-shaped regions (indicated by white arrows). Bacteria were visualized with a 100× objective.
Figure 6. M. smegmatis ponA1 complements ponA1 operon depletion.
(A) The M. smegmatis ponA1 depletion strain was complemented with either a vector expressing an operon of M. smegmatis ponA1 and red fluorescent protein (RFP) or RFP alone. Complemented strains were grown in the presence (50 ng/ml ATC) or absence of inducer and growth was determined by OD600. While the RFP complemented strain only grew in the presence of inducer, the PBP1 complemented strain grew normally in both the absence and presence of inducer. (B) At 36 hours post-induction, cultures from RFP and ponA1 complemented strains were photographed. Data is representative of several biological replicates for both strains. (C) Microscopic analysis of complemented strains—either the RFP complemented strain grown with inducer or the ponA1 complemented strain without inducer—showed that both strains expressed RFP from the complementation vector and appear morphologically normal.
Figure 7. PBP1 inhibits the synergistic hydrolysis of cell wall by RipA-RpfB complex.
N-terminal GST fusion proteins were expressed and purified from E. coli. Equal amounts of individual or combinations of proteins were incubated with insoluble FITC-labeled substrate: M. luteus cell wall material (A) or Streptomyces peptidoglycan (B). The extent of hydrolysis was determined by measuring the amount of soluble FITC remaining after centrifugation, and thus released during hydrolysis of the insoluble substrate. GST and buffer alone were used to determine background release of FITC and were subtracted from final values. Data are from representative experiments, each done in triplicate. Data are represented as mean +/− SEM. p-values for one-tailed, unpaired t-tests: (A) 1 vs. 2: 0.027 significant, 2 vs. 4: 0.016 significant, 1 vs. 3: 0.074 not significant, 1 vs. 4: 0.188 not significant, (B): 1 vs. 2: 0.009 significant, 2 vs. 4: 0.018 significant, 1 vs. 3: 0.279 not significant, 1 vs. 4: 0.240 not significant (significant to p<0.05). (C) Schematic diagram of the basic structure of mycobacterial peptidoglycan, indicating where RpfB and RipA are predicted to have hydrolytic activity and where PBP1 is predicted to have synthetic activity. Lines connecting NAG to NGM represent β-1,4-glycosidic bonds, while lines connecting NGM to NGM represent peptide cross-linkages. NAG: N-acetyl glucosamine, NGM: N-glycolyl muramic acid, NAM: N-acetyl muramic acid (note that mycobacteria have a mixture of NGM and NAM, with the NGM structure shown here).
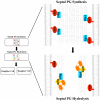
Figure 8. Model of RipA regulation by PBP1 during septation.
A RipA-PBP1 complex that exists at the initiation of septation may synthesize a thick layer of peptidoglycan (PG) between daughter cells during the process of cytokinesis. After PG synthesis and fission are finished, RipA may exchange PBP1 for autolysin binding partners like RpfB. These new complexes are highly efficient at PG hydrolysis and will separate the cell walls of the two mature daughter cells.
Similar articles
- A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor.
Hett EC, Chao MC, Deng LL, Rubin EJ. Hett EC, et al. PLoS Pathog. 2008 Feb 29;4(2):e1000001. doi: 10.1371/journal.ppat.1000001. PLoS Pathog. 2008. PMID: 18463693 Free PMC article. - A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis.
Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. Hett EC, et al. Mol Microbiol. 2007 Nov;66(3):658-68. doi: 10.1111/j.1365-2958.2007.05945.x. Epub 2007 Oct 4. Mol Microbiol. 2007. PMID: 17919286 - Wake up! Peptidoglycan lysis and bacterial non-growth states.
Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. Keep NH, et al. Trends Microbiol. 2006 Jun;14(6):271-6. doi: 10.1016/j.tim.2006.04.003. Epub 2006 May 3. Trends Microbiol. 2006. PMID: 16675219 Review. - The dynamic region of the peptidoglycan synthase gene, Rv0050, induces the growth rate and morphologic heterogeneity in Mycobacteria.
Gao B, Wang J, Huang J, Huang X, Sha W, Qin L. Gao B, et al. Infect Genet Evol. 2019 Aug;72:86-92. doi: 10.1016/j.meegid.2018.12.012. Epub 2018 Dec 10. Infect Genet Evol. 2019. PMID: 30543940 - The Cell Wall Hydrolytic NlpC/P60 Endopeptidases in Mycobacterial Cytokinesis: A Structural Perspective.
Squeglia F, Moreira M, Ruggiero A, Berisio R. Squeglia F, et al. Cells. 2019 Jun 18;8(6):609. doi: 10.3390/cells8060609. Cells. 2019. PMID: 31216697 Free PMC article. Review.
Cited by
- The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan.
Alderwick LJ, Harrison J, Lloyd GS, Birch HL. Alderwick LJ, et al. Cold Spring Harb Perspect Med. 2015 Mar 27;5(8):a021113. doi: 10.1101/cshperspect.a021113. Cold Spring Harb Perspect Med. 2015. PMID: 25818664 Free PMC article. Review. - Lytic transglycosylases: concinnity in concision of the bacterial cell wall.
Dik DA, Marous DR, Fisher JF, Mobashery S. Dik DA, et al. Crit Rev Biochem Mol Biol. 2017 Oct;52(5):503-542. doi: 10.1080/10409238.2017.1337705. Epub 2017 Jun 23. Crit Rev Biochem Mol Biol. 2017. PMID: 28644060 Free PMC article. Review. - FtsEX-independent control of RipA-mediated cell separation in Corynebacteriales.
Gaday Q, Megrian D, Carloni G, Martinez M, Sokolova B, Ben Assaya M, Legrand P, Brûlé S, Haouz A, Wehenkel AM, Alzari PM. Gaday Q, et al. Proc Natl Acad Sci U S A. 2022 Dec 13;119(50):e2214599119. doi: 10.1073/pnas.2214599119. Epub 2022 Dec 5. Proc Natl Acad Sci U S A. 2022. PMID: 36469781 Free PMC article. - Bacterial cell division regulation by Ser/Thr kinases: a structural perspective.
Ruggiero A, De Simone P, Smaldone G, Squeglia F, Berisio R. Ruggiero A, et al. Curr Protein Pept Sci. 2012 Dec;13(8):756-66. doi: 10.2174/138920312804871201. Curr Protein Pept Sci. 2012. PMID: 23305362 Free PMC article. Review. - A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria.
Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei JR, Dhiman RK, Crick DC, Rubin EJ, Sassetti CM, Alber T. Gee CL, et al. Sci Signal. 2012 Jan 24;5(208):ra7. doi: 10.1126/scisignal.2002525. Sci Signal. 2012. PMID: 22275220 Free PMC article.
References
- Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. - PubMed
- Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. - PubMed
- Doyle RJ, Marquis RE. Elastic, flexible peptidoglycan and bacterial cell wall properties. Trends Microbiol. 1994;2:57–60. - PubMed
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI048704/AI/NIAID NIH HHS/United States
- R01 AI051929/AI/NIAID NIH HHS/United States
- R01 AI48704/AI/NIAID NIH HHS/United States
- P01 AI056296/AI/NIAID NIH HHS/United States
- R01 AI51929/AI/NIAID NIH HHS/United States
- P01 AI56296/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials