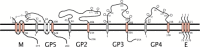The structural biology of PRRSV - PubMed (original) (raw)
Review
The structural biology of PRRSV
Terje Dokland. Virus Res. 2010 Dec.
Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is an enveloped, positive-sense single-stranded RNA virus belonging to the Arteriviridae family. Arteriviruses and coronaviruses are grouped together in the order Nidovirales, based on similarities in genome organization and expression strategy. Over the past decade, crystal structures of several viral proteins, electron microscopic studies of the virion, as well as biochemical and in vivo studies on protein-protein interactions have led to a greatly increased understanding of PRRSV structural biology. At this point, crystal structures are available for the viral proteases NSP1α, NSP1β and NSP4 and the nucleocapsid protein, N. The NSP1α and NSP1β structures have revealed additional non-protease domains that may be involved in modulation of host functions. The N protein forms a dimer with a novel fold so far only seen in PRRSV and other nidoviruses. Cryo-electron tomographic studies have shown the three-dimensional organization of the PRRSV virion and suggest that the viral nucleocapsid has an asymmetric, linear arrangement, rather than the isometric core previously described. Together, these studies have revealed a closer structural relationship between arteri- and coronaviruses than previously anticipated.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures
Fig. 1
Ribbon diagrams of the crystal structures of PRRSV NSP1α (A; PDB accession code 3IFU), NSP1β (B; 3MTV) and NSP4 (C; 3FAN) (Sun et al., 2009, Tian et al., 2009, Xue et al., 2010). In (A), the N-terminal zinc finger domain of NSP1α is colored green, the protease domain is blue. The active site residues Cys 76 and His 146 as well as the zinc finger Cys residues (Cys 8, Cys 20, Cys 25 and Cys 28) are shown in stick representation, and the two zinc ions are shown as pink balls. In (B), the N-terminal nuclease domain of NSP1β (including the linker between the two domains) is shown in yellow, while the protease domain is blue. The active site residues Cys 90 and His 159 are shown in stick representation. In (C), the chymotrypsin-like domain of NSP4 is blue, while the C-terminal domain is red. The active site residues His 39, Asp 64 and Ser 118 are shown as sticks. The figure was made with UCSF Chimera (Pettersen et al., 2004). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2
Topology of PRRSV envelope proteins. Residue numbering is according to VR-2332 (a type 1 strain). Transmembrane domains are shown as rectangles crossing the lipid bilayer. The stippled boxes represent predicted signal peptides; the cleavage site is indicated by the broken line. (The GP3 signal peptide may not get cleaved.) Glycosylation is indicated by hexagons with the corresponding residues numbered. The disulfide link between M and GP5 is also indicated. E forms homo-oligomers and is shown as a trimer, but the exact number is not known.
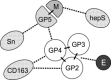
Fig. 3
Map of protein–protein interactions in the PRRSV envelope. The viral proteins are indicated as circles. Solid lines indicate the well-established, possibly covalent interactions that define the two major envelope protein complexes M–GP5 and GP2–GP3–GP4. Dashed lines indicate other (non-covalent) interactions. Interactions with the three host receptors proteins heparan sulfate (hepS), sialoadhesin (Sn) and CD163 are also shown. E interacts with the GP2–GP3–GP4 complex, but it is unclear which specific protein that is involved.
Fig. 4
Structure of the nucleocapsid protein. (A) Sequence of N, showing the division into an N-terminal RNA-binding domain (residues 1–57) and a C-terminal dimerization domain (58–123), represented by the NΔ57 construct. Secondary structure elements are shown, including the predicted hydrophobic helix (α0) in the RNA-binding domain. Positively charged residues in the RNA-binding domain are indicated by (+) signs. (B and C) Ribbon diagrams of the NΔ57 dimer (PDB accession code 1P65) in the “top” view (B), looking down on the two long α2-helices, and a “side” view (C), looking approximately perpendicular to the two α2-helices (Doan and Dokland, 2003). Positions of N and C termini and the extension of the N-terminal RNA-binding domains are indicated.
Fig. 5
Representative examples of PRRSV virions imaged by (A) negative stain EM (stained with 1% uranyl acetate) and (B) by cryo-EM. The inset in (B) shows one typical particle with pertinent dimensions indicated. Scale bar, 100 nm.
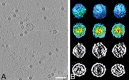
Fig. 6
Cryo-electron tomography of PRRSV. (A) Central section (15.8 Å thick) through one tomogram, showing numerous PRRSV virions. Scale bar, 200 nm. (B) Montage of individual PRRSV sub-tomograms. Each column represents a distinct particle in various representations, as follows: top row, isosurface of whole particle, viewed along _y_-axis (“side view”); second row, isosurface of half particle, colored from red to blue according to distance from the center; third row, central section through particles along _y_-axis (“side view”, viewed perpendicular to the direction of the electron beam); bottom row, central section along _z_-axis (“top view”, viewed parallel to the direction of the beam) (Spilman et al., 2009). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7
Possible model for the nucleocapsid organization in the virion. The stippled oval shapes represent the N protein dimers. Pertinent structural features are indicated, including the α0, α2 and α3-helices and the positively charged N-terminal domains. These dimers are organized in a roughly helical fashion around the RNA (grey sinusoidal curve) through interactions with the N-terminal RNA-binding domains. This N–RNA ribbon is folded into the nucleocapsid, giving rise to the double-layered appearance of the viral core in electron micrographs, as shown. The “train track” represents the viral envelope with the TM domains of the major envelope proteins.
Similar articles
- Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha.
Sun Y, Xue F, Guo Y, Ma M, Hao N, Zhang XC, Lou Z, Li X, Rao Z. Sun Y, et al. J Virol. 2009 Nov;83(21):10931-40. doi: 10.1128/JVI.02579-08. Epub 2009 Aug 12. J Virol. 2009. PMID: 19706710 Free PMC article. - A Dimerization-Dependent Mechanism Drives the Endoribonuclease Function of Porcine Reproductive and Respiratory Syndrome Virus nsp11.
Shi Y, Li Y, Lei Y, Ye G, Shen Z, Sun L, Luo R, Wang D, Fu ZF, Xiao S, Peng G. Shi Y, et al. J Virol. 2016 Apr 14;90(9):4579-4592. doi: 10.1128/JVI.03065-15. Print 2016 May. J Virol. 2016. PMID: 26912626 Free PMC article. - Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid.
Spilman MS, Welbon C, Nelson E, Dokland T. Spilman MS, et al. J Gen Virol. 2009 Mar;90(Pt 3):527-535. doi: 10.1099/vir.0.007674-0. J Gen Virol. 2009. PMID: 19218197 - Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates.
Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Dea S, et al. Arch Virol. 2000;145(4):659-88. doi: 10.1007/s007050050662. Arch Virol. 2000. PMID: 10893147 Free PMC article. Review. - The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis.
Music N, Gagnon CA. Music N, et al. Anim Health Res Rev. 2010 Dec;11(2):135-63. doi: 10.1017/S1466252310000034. Epub 2010 Apr 14. Anim Health Res Rev. 2010. PMID: 20388230 Review.
Cited by
- Identification of an Intramolecular Switch That Controls the Interaction of Helicase nsp10 with Membrane-Associated nsp12 of Porcine Reproductive and Respiratory Syndrome Virus.
Hu Y, Ke P, Gao P, Zhang Y, Zhou L, Ge X, Guo X, Han J, Yang H. Hu Y, et al. J Virol. 2021 Aug 10;95(17):e0051821. doi: 10.1128/JVI.00518-21. Epub 2021 Aug 10. J Virol. 2021. PMID: 34076477 Free PMC article. - Disulfide linkages mediating nucleocapsid protein dimerization are not required for porcine arterivirus infectivity.
Zhang R, Chen C, Sun Z, Tan F, Zhuang J, Tian D, Tong G, Yuan S. Zhang R, et al. J Virol. 2012 Apr;86(8):4670-81. doi: 10.1128/JVI.06709-11. Epub 2012 Feb 1. J Virol. 2012. PMID: 22301142 Free PMC article. - Point-of-Care and Label-Free Detection of Porcine Reproductive and Respiratory Syndrome and Swine Influenza Viruses Using a Microfluidic Device with Photonic Integrated Circuits.
Manessis G, Frant M, Wozniakowski G, Nannucci L, Benedetti M, Denes L, Gyula B, Gelasakis AI, Squires C, Recuero S, Sanchez C, Griol A, Giusti A, Bossis I. Manessis G, et al. Viruses. 2022 May 7;14(5):988. doi: 10.3390/v14050988. Viruses. 2022. PMID: 35632730 Free PMC article. - Porcine Reproductive and Respiratory Syndrome Virus nsp11 Antagonizes Type I Interferon Signaling by Targeting IRF9.
Wang D, Chen J, Yu C, Zhu X, Xu S, Fang L, Xiao S. Wang D, et al. J Virol. 2019 Jul 17;93(15):e00623-19. doi: 10.1128/JVI.00623-19. Print 2019 Aug 1. J Virol. 2019. PMID: 31092569 Free PMC article. - Pathological observation and transcriptomic analysis of thymus injury in PRRSV-infected piglets.
Su N, Lin Z, Liu X, Sun X, Jin X, Feng H, Zhan C, Hu X, Gu C, Zhang W, Cheng G. Su N, et al. Vet Res Commun. 2023 Dec;47(4):1949-1962. doi: 10.1007/s11259-023-10133-x. Epub 2023 Jun 2. Vet Res Commun. 2023. PMID: 37266866 Free PMC article.
References
- Allaire M., Chernala M.M., Malcolm B.A., James M.N.G. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like proteinases. Nature. 1994;369:72–76. - PubMed
- Allende R., Lewis T.L., Lu Z., Rock D.L., Kutish G.F., Ali A., Doster A.R., Osorio F.A. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 1999;80:307–315. - PubMed
- Balasuriya U.B., MacLachlan N.J. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 2004;102:107–129. - PubMed
- Barrette-Ng I.H., Ng K.K.S., Mark B.L., van Aken D., Cherney M.M., Garen C., Kolodenko Y., Gorbalenya A.E., Snijder E.J., James M.N.G. Structure of arterivirus nsp4. J. Biol. Chem. 2002;277:39960–39966. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous