The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor - PubMed (original) (raw)
The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor
Yi-Chun Yeh et al. J Bacteriol. 2010 Oct.
Abstract
Cell division in Caulobacter crescentus involves constriction and fission of the inner membrane (IM) followed about 20 min later by fission of the outer membrane (OM) and daughter cell separation. In contrast to Escherichia coli, the Caulobacter Tol-Pal complex is essential. Cryo-electron microscopy images of the Caulobacter cell envelope exhibited outer membrane disruption, and cells failed to complete cell division in TolA, TolB, or Pal mutant strains. In wild-type cells, components of the Tol-Pal complex localize to the division plane in early predivisional cells and remain predominantly at the new pole of swarmer and stalked progeny upon completion of division. The Tol-Pal complex is required to maintain the position of the transmembrane TipN polar marker, and indirectly the PleC histidine kinase, at the cell pole, but it is not required for the polar maintenance of other transmembrane and membrane-associated polar proteins tested. Coimmunoprecipitation experiments show that both TolA and Pal interact directly or indirectly with TipN. We propose that disruption of the trans-envelope Tol-Pal complex releases TipN from its subcellular position. The Caulobacter Tol-Pal complex is thus a key component of cell envelope structure and function, mediating OM constriction at the final step of cell division as well as the positioning of a protein localization factor.
Figures
FIG. 1.
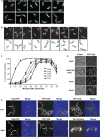
The Caulobacter crescentus tol-pal gene cluster. (A) Predicted organization of the components of the Tol-Pal complex in the E. coli (35) and Caulobacter cell envelopes. TolQ, TolR, and TolA are integral IM proteins, and Pal is an OM protein that interacts with the periplasmic TolB protein. The Caulobacter TipN polar marker is an IM protein (25, 29). (B) Schematic of the gene organization of the Caulobacter tol-pal components. Arrows indicate the direction of transcription and putative position of promoters. The +1 transcriptional start site is at −49 of the tolQ coding sequence, as indicated (36). (C) mRNA expression patterns of genes encoding the components of the Tol-Pal complex over the course of a cell cycle. pal expression peaks in the swarmer cell and drops thereafter, while expression of the other genes is not cell cycle dependent. (D) Western blot analysis of relative Pal protein levels (upper panel; arrows indicate the Pal protein) during the cell cycle starting with a synchronous population of wild-type swarmer cells. The FtsZ protein (lower panel) is shown as a loading control and quality control for the synchrony. (E) Normalized abundance of Pal and FtsZ protein levels over the course of the cell cycle. The Pal protein level does not exhibit significant changes during the course of the cell cycle.
FIG. 2.
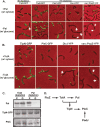
Subcellular localization of the proteins encoded by the tol-pal cluster. (A) Fusions of TolQ, TolR, TolA, TolB, and Pal to GFP, YFP, or mCherry, at either N or C terminals, were generated and their cellular locations were observed by fluorescence microscopy. LS4517 (P_xylX_-tolQ-yfp), LS4518 (P_xylX_-gfp-tolR), LS4519 (P_xylX_-gfp-tolA), LS4520 (P_xylX_-mCherry-tolB), and LS4521 (P_xylX_-pal-mCherry) strains were each incubated in the presence of 0.3% xylose for 2 h to induce the expression of the fluorescently tagged fusions in a merodiploid cell containing the wild-type gene. All of the fusion proteins tested localized to both the cell pole (upper panel) and to the division plane (lower panel). (B) A synchronized LS4519 swarmer cell population, producing GFP-TolA, was suspended in PYE medium containing 0.3% xylose and allowed to proceed through the cell cycle. Samples were visualized at 20-min intervals by phase-contrast and fluorescence microscopy. A schematic of Caulobacte_r cells showing the dynamic localization of TolA as a function of the cell cycle is shown below the micrographs. The accumulation of GFP-TolA at the division plane is indicated by arrows. (C) Cells of strains EG052 (PxylX-ftsZ-venus), EG055 (PxylX-venus_-ftsA), EG051 (PxylX-venus_-ftsI), LS4517 (PxylX-tolQ-venus), and LS4519 (PxylX-gfp_-tolA) were grown to exponential phase in PYE medium, induced for 1 h with 0.3% xylose, synchronized, and resuspended in PYE medium supplemented with 0.3% xylose. Cells were withdrawn from the cultures at 10-min intervals and visualized by phase-contrast and fluorescence microscopy to determine if the fluorescently labeled proteins were positioned at the incipient division plane. At least 150 cells were analyzed per time point. (D) Localization of YFP-TolA in strain LS4546 depleted of FtsZ for 3 h (−FtsZ) and when FtsZ was depleted for 3 h and then was FtsZ added back by incubation in the presence of the xylose inducer for 0.5 h. In both cases, cultures were incubated in the presence of 0.5 mM vanillate to induce the expression of y_fp-tolA. Localization of YFP-TolA in strain LS4547 depleted of FtsA after growth in PYE medium with glucose (PYEG) for 8 h is shown. White arrows indicate foci of YFP-TolA. Localization of YFP-TolA in strain LS4626 (xylX::P_xylX-yfp-tolA Δ_ftsN vanA::P_van-ftsN) depleted of FtsN after growth in PYE medium without vanillate for 14 h is also shown. Two hours before analysis, expression of YFP-TolA was induced by addition of 0.3% xylose. YFP-TolA requires FtsZ, but not FtsA or FtsN, to form fluorescent foci. (E) Fluorescence microscopy of Pal depletion strains carrying plasmid-borne P_van_-tolQ-yfp, P_van-yfp-tolR_, or P_van-yfp-tolA_ incubated in the absence of xylose (−Pal). Cells were grown in PYEX, washed, and incubated in PYEG for 11 h to deplete Pal. TolA depletion strains bearing the plasmid P_van_-tolQ-yfp or P_van-yfp-tolR_ were incubated in the absence of xylose (−TolA) for 11 h. A TolA depletion strain, with P_van-pal-mCherry_ integrated at the chromosomal vanA locus, was incubated in the presence of 0.5 mM vanillate to induce Pal-mCherry and in the absence of xylose for 11 h (−TolA). Cells were imaged by using phase-contrast and fluorescence microscopy.
FIG. 3.
Mislocalization of the TipN-GFP and PleC-GFP polar proteins in the absence of the Pal or TolA proteins. (A) Fluorescence microscopy of Pal depletion strains bearing TipN-GFP, PleC-GFP, DivJ-YFP, or PopZ-YFP grown in the presence of xylose to induce Pal accumulation or in the presence of glucose to deplete Pal. Strain LS4528 (tipN-gfp Δ_pal_ P_xylX-pal_) was imaged after either 9 or 11 h of incubation in the absence of xylose to deplete Pal. The upper panels show the subcellular location of TipN-GFP, PleC-GFP, and DivJ-YFP in cells grown in the presence of xylose to induce Pal, compared to cells shown in the lower panels incubated with glucose to deplete Pal. Strain LS4534 (Δ_pal_ P_xylX-pal_ P_van-popZ-yfp_) was incubated for 1 h in the presence of 0.5 mM vanillate to induce PopZ-YFP in the presence of either xylose (+Pal) or glucose (−Pal). (B) The same fusion proteins were examined by fluorescence microscopy in TolA depletion strains LS4529 (tipN-gfp Δ_tolA_ P_xylX-tolA_), LS4531 (pleC-gfp Δ_tolA_ P_xylX-tolA_), LS4533 (divJ-yfp Δ_tolA_ P_xylX-tolA_), and LS4535 (Δ_tolA_ P_xylX-tolA_ P_van-popZ-yfp_) incubated for 11 h in the presence of either xylose (+TolA) or glucose (−TolA). Arrows indicate polar localization of DivJ-YFP and PopZ-YFP in the absence of either Pal or TolA. (C) Immunoblot analysis of Pal, TipN-GFP, and PleC levels in cell extracts of TolA depletion and Pal depletion cultures. Neither TipN-GFP nor PleC levels were affected after 11 h of growth in the absence of xylose. (D) The predicted localization dependency pathway derived from experiments shown in this figure and in Fig. 2.
FIG. 4.
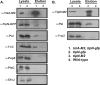
The Tol-Pal complex interacts with TipN-GFP. (A) In vivo coimmunoprecipitation of TolA-M2 in strain LS4536 (tipN-gfp Δ_tolA_ P_xylX-tolA-m2_) and LS4529 (tipN-gfp Δ_tolA_ P_xylX-tolA_). Western blots of whole-cell extracts (lysate) and eluted samples were probed with the indicated antibodies (anti-M2 to probe TolA-M2 and anti-GFP to probe TipN-GFP, anti-Pal, anti-FtsZ, anti-DivJ, anti-PleC, and anti-PopZ). TipN, FtsZ, and Pal, but not DivJ, PleC, or PopZ, were observed to interact, directly or indirectly, with TolA-M2. (B) Coimmunoprecipitation of Pal with TipN-M2 in strain LS4538 (P_xylX-tipN-m2_) and in wild-type cells. Western blots of eluted samples were probed with anti-M2 to probe TipN-M2, anti-Pal, and anti-PopZ. Pal but not PopZ was observed to interact with TipN-M2.
FIG. 5.
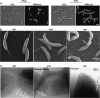
Phenotypes of Pal depletion strains. (A) To visualize cell morphology in cells depleted of Pal (right panels), compared to cells containing Pal (left panels), strain LS4524 was grown in PYEX to induce pal expression. Cells were then washed and grown in PYEG for 10 h to deplete Pal. Cells grown in either PYEX or PYEG were stained with FM4-64 and visualized by light microscopy. Bar, 2 μm. When strain LS4524 was grown in PYEG for 10 h, the cells exhibited a late-stage cell division defect and polar blebs (white arrows). (B) Scanning EM images of the Pal depletion strain LS4524 incubated in the presence of either xylose or glucose for 12 h. Wild-type cells and strain LS4524 grown in the presence of xylose to induce expression of Pal were similar. However, in the absence of Pal, surface blebs were visible across the lateral cell surface, at the cell poles, and at the division plane (indicated by arrows). (C) A cryo-EM image of wild-type cells exhibiting a well-defined IM, PG layer, OM, and S-layer. Visualization of strain LS4524 after Pal depletion by growth in PYEG for 10 h showed aberrant OM structures at the cell pole and at the site of division.
FIG. 6.
Phenotype of TolA and TolB depletion strains. (A) SEM images of the TolA depletion strain LS4525 grown in the presence of either xylose or glucose for 12 h (to deplete TolA). (B) SEM images of the TolB depletion strain LS4526 grown for 12 h in the presence of either xylose or glucose (to deplete TolB). Depletion of either TolA or TolB yielded cells with extensive surface blebs. Bar (A and B), 1 μm. (C) Cryo-EM images of strain LS4525 grown for 11 h in glucose to deplete TolA, showing significant OM disruptions both laterally and at the cell poles (arrows). (D) Cryo-EM images of strain LS4526 grown for 11 h in glucose to deplete TolB show OM defects at both the cell poles and the division plane (arrows). Bar (C and D,) 100 nm.
FIG. 7.
Structural relationship of the peptidoglycan layer and the IM and OM in TolA and Pal depletion strains. (A) Cryo-EM of strain LS4524 after Pal depletion by growth in PYEG for 10 h showed an OM polar bleb with the peptidoglycan layer (arrows) dissociated from the OM but adhering to the IM. (B) Cryo-EM of strain LS4525 grown for 11 h in glucose to deplete TolA showed an OM polar bleb with the peptidoglycan layer (arrows) adhering to the OM but separated from the IM. (C) Lateral cell envelope of strain LS4524 depleted of Pal with the OM separated from the peptidoglycan layer (arrows). (D) Lateral cell envelope of strain LS4525 depleted of TolA, with the peptidoglycan layer separated from the IM.
Similar articles
- DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter.
Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. Goley ED, et al. Mol Microbiol. 2010 Jul 1;77(1):56-73. doi: 10.1111/j.1365-2958.2010.07222.x. Epub 2010 May 24. Mol Microbiol. 2010. PMID: 20497504 Free PMC article. - Timing of TolA and TolQ Recruitment at the Septum Depends on the Functionality of the Tol-Pal System.
Baccelli P, Rachedi R, Serrano B, Petiti M, Bernard CS, Houot L, Duche D. Baccelli P, et al. J Mol Biol. 2022 Apr 15;434(7):167519. doi: 10.1016/j.jmb.2022.167519. Epub 2022 Feb 28. J Mol Biol. 2022. PMID: 35240126 - Loss of Bacterial Cell Pole Stabilization in Caulobacter crescentus Sensitizes to Outer Membrane Stress and Peptidoglycan-Directed Antibiotics.
Vallet SU, Hansen LH, Bistrup FC, Laursen SA, Chapalay JB, Chambon M, Turcatti G, Viollier PH, Kirkpatrick CL. Vallet SU, et al. mBio. 2020 May 5;11(3):e00538-20. doi: 10.1128/mBio.00538-20. mBio. 2020. PMID: 32371598 Free PMC article. - Force-Generation by the Trans-Envelope Tol-Pal System.
Webby MN, Williams-Jones DP, Press C, Kleanthous C. Webby MN, et al. Front Microbiol. 2022 Mar 3;13:852176. doi: 10.3389/fmicb.2022.852176. eCollection 2022. Front Microbiol. 2022. PMID: 35308353 Free PMC article. Review. - A new factor stimulating peptidoglycan hydrolysis to separate daughter cells in Caulobacter crescentus.
Collier J. Collier J. Mol Microbiol. 2010 Jul 1;77(1):11-4. doi: 10.1111/j.1365-2958.2010.07225.x. Epub 2010 May 24. Mol Microbiol. 2010. PMID: 20497501 Review.
Cited by
- Gram-Negative Bacteria's Outer Membrane Vesicles.
Kim JY, Suh JW, Kang JS, Kim SB, Yoon YK, Sohn JW. Kim JY, et al. Infect Chemother. 2023 Mar;55(1):1-9. doi: 10.3947/ic.2022.0145. Epub 2023 Jan 6. Infect Chemother. 2023. PMID: 36731499 Free PMC article. Review. - Helicobacter pylori HP0135 Is a Small Lipoprotein That Has a Role in Outer Membrane Stability.
Nguyen D, Ivester RG, Rosinke K, Hoover TR. Nguyen D, et al. Molecules. 2025 Jan 7;30(2):204. doi: 10.3390/molecules30020204. Molecules. 2025. PMID: 39860075 Free PMC article. - The multifarious roles of Tol-Pal in Gram-negative bacteria.
Szczepaniak J, Press C, Kleanthous C. Szczepaniak J, et al. FEMS Microbiol Rev. 2020 Jul 1;44(4):490-506. doi: 10.1093/femsre/fuaa018. FEMS Microbiol Rev. 2020. PMID: 32472934 Free PMC article. Review. - A polarity factor takes the lead in chromosome segregation.
Kirkpatrick CL, Viollier PH. Kirkpatrick CL, et al. EMBO J. 2010 Sep 15;29(18):3035-6. doi: 10.1038/emboj.2010.213. EMBO J. 2010. PMID: 20842173 Free PMC article. - Bacterial Outer Membrane Vesicles: From Discovery to Applications.
Sartorio MG, Pardue EJ, Feldman MF, Haurat MF. Sartorio MG, et al. Annu Rev Microbiol. 2021 Oct 8;75:609-630. doi: 10.1146/annurev-micro-052821-031444. Epub 2021 Aug 5. Annu Rev Microbiol. 2021. PMID: 34351789 Free PMC article. Review.
References
- Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. - PubMed
- Anwari, K., S. Poggio, A. Perry, X. Gatsos, S. H. Ramarathinam, N. A. Williamson, N. Noinaj, S. Buchanan, K. Gabriel, A. W. Purcell, C. Jacobs-Wagner, and T. Lithgow. 2010. A modular BAM complex in the outer membrane of the alpha-proteobacterium Caulobacter crescentus. PLoS One 5:e8619. - PMC - PubMed
- Bouveret, E., R. Derouiche, A. Rigal, R. Lloubes, C. Lazdunski, and H. Benedetti. 1995. Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J. Biol. Chem. 270:11071-11077. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous