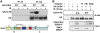Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4 - PubMed (original) (raw)
Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4
Sophie E Polo et al. EMBO J. 2010.
Abstract
The chromatin remodelling factor chromodomain helicase DNA-binding protein 4 (CHD4) is a catalytic subunit of the NuRD transcriptional repressor complex. Here, we reveal novel functions for CHD4 in the DNA-damage response (DDR) and cell-cycle control. We show that CHD4 mediates rapid poly(ADP-ribose)-dependent recruitment of the NuRD complex to DNA-damage sites, and we identify CHD4 as a phosphorylation target for the apical DDR kinase ataxia-telangiectasia mutated. Functionally, we show that CHD4 promotes repair of DNA double-strand breaks and cell survival after DNA damage. In addition, we show that CHD4 acts as an important regulator of the G1/S cell-cycle transition by controlling p53 deacetylation. These results provide new insights into how the chromatin remodelling complex NuRD contributes to maintaining genome stability.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
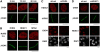
CHD4 is recruited to sites of laser-induced DNA damage within the NuRD complex. (A) Immunodetection of CHD4 and γH2AX (damage sites) at the indicated times after laser micro-irradiation in U2OS cells. (B) Recruitment of NuRD complex subunits to sites of laser-induced DNA damage (labelled by γH2AX) 5 min after micro-irradiation in U2OS cells. (C, D) Immunodetection of HDAC1 and CHD4 5 min after laser micro-irradiation in U2OS cells treated with the indicated siRNAs (siLuci: control). Cells were pre-treated with ATM inhibitor to facilitate detection of HDAC1 and CHD4 lines. Lower panels show siRNA efficiency. In laser micro-irradiation experiments, detergent pre-extraction was performed before fixation of the cells for immunostaining. Scale bars, 10 μm.
Figure 2
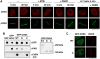
PARP-dependent recruitment of CHD4 to sites of DNA damage. (A) Immunodetection of CHD4 and γH2AX (damage sites) at the indicated times after laser micro-irradiation in U2OS (left) and A-T cells (right). Cells were pre-incubated for 1 h with the indicated inhibitors (ATMi, ATM inhibitor; PARPi, PARP inhibitor) before micro-irradiation. (B) PAR-binding assay with immunoprecipitated GFP-CHD4 wild-type (WT: residues 1–1937) or truncated mutants (N: residues 1–758; C: residues 1183–1937). GFP only is used as a negative control and GFP-APLF as a positive control. Right panel: corresponding Ponceau staining of the blot after immunoprecipitation from HEK-293 cells. (C) Recruitment of GFP-CHD4 wild-type (WT) or a truncated mutant (C: residues 1183–1937) to sites of laser-induced damage (γH2AX) 5 min after micro-irradiation in U2OS cells. In all cases, detergent pre-extraction was performed before fixation of the cells for immunostaining. Scale bars, 10 μm.
Figure 3
ATM-dependent phosphorylation of CHD4 Ser-1346 in response to DNA damage. (A) Detection of CHD4 phosphorylation (αphos-Ab) 1 h after exposure to 10 Gy ionizing radiation (+ IR) on HA immunoprecipitates from U2OS cells transiently expressing HA-CHD4 wild-type (WT) or S1346A point mutant (SA). The scheme represents CHD4 protein with the amino-acid position of the candidate phospho-serine (S1346); the N and C fragments correspond to CHD4 truncated mutants analysed in Figures 2 and 4. Domain organization of CHD4: NLS (putative nuclear localization signal), PHD (plant homeodomain), CHROMO (chromodomain), DEXH (ATP-binding domain), HELIC (helicase carboxyl-terminal domain). (B) Detection of CHD4 S1346 phosphorylation (αphos-Ab) 30 min after cell exposure to 10 Gy ionizing radiation (IR) on HA immunoprecipitates from U2OS cells transiently expressing HA-CHD4. Cells were pre-treated with the indicated inhibitors (W, Wortmannin; PKi, DNA-PK inhibitor; ATMi, ATM inhibitor). ATM-dependent phosphorylation of SMC1 S966 and DNA-PKcs autophosphorylation on S2056 are used as controls.
Figure 4
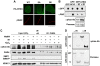
DNA-damage-induced phosphorylation and recruitment of CHD4 to DNA lesions are distinct events. (A) Recruitment of HA-CHD4 wild-type (WT) and Ser-1346 point mutants (SA, mutated to Ala; SE, mutated to Glu) to sites of laser-induced damage (γH2AX) in transiently transfected U2OS cells 5 min after micro-irradiation. Scale bar, 10 μm. (B) PAR-binding activity of GFP-CHD4 from HEK-293 cells exposed or not to ionizing radiation (IR). GFP only is used as a negative control, GFP-APLF as a positive control. Lower panel shows CHD4 phosphorylation after IR analysed in parallel by western blotting. (C) Detection of CHD4 phosphorylation (αphos-Ab) 30 min after exposure to 10 Gy ionizing radiation (IR) or 500 μM hydrogen peroxide (H2O2) on CHD4 immunoprecipitates from HEK-293 cells treated or not with PARP inhibitor (PARPi). SMC1 phosphorylation is used as a control for DNA damage. The NuRD subunit HDAC1 co-immunoprecipitates with CHD4. (D) Detection of CHD4 phosphorylation (αphos-Ab) 30 min after exposure to 10 Gy ionizing radiation (IR) on GFP immunoprecipitates from H3K293 cells transiently expressing GFP-CHD4 wild-type (WT) or a truncated mutant (C: residues 1183–1937).
Figure 5
CHD4 regulates cell sensitivity to DNA damage. (A) γH2AX foci formation in U2OS cells upon CHD4 depletion (siCHD4) compared with control (siLuci) 1 h after exposure to 10 Gy ionizing radiation (IR). (B) Phosphorylation of DNA-damage checkpoint proteins analysed by western blotting of total extracts prepared from HeLa cells at the indicated times after cell exposure to 10 Gy ionizing radiation (IR) (siCHD4, CHD4 depletion; siLuci, control). (C) Recruitment of MDC1, 53BP1 and BRCA1 to sites of laser-induced damage (γH2AX) 5–10 min after micro-irradiation in HeLa cells upon CHD4 depletion (siCHD4) compared with control (siLuci). Detergent pre-extraction was performed before fixation and immunostaining for BRCA1. Scale bars, 10 μm. (D) Efficiency of DSB repair in control (siLuci) or CHD4-depleted U2OS cells analysed by neutral comet assay after phleomycin treatment (Phleo). Error bars: s.d. from two independent experiments. (E) Clonogenic survival of U2OS cells upon CHD4 knock-down (siCHD4) compared with control (siLuci) in response to ionizing radiation (IR) or hydrogen peroxide (H2O2). H2O2 treatment was for 10 min at the indicated doses. Error bars indicate s.d. from two and three independent experiments, respectively. The results are normalized to plating efficiencies to focus on the effect of CHD4 depletion upon DNA damage. Note that in the absence of DNA-damaging agent, the viability of CHD4-depleted cells is ∼66% of that of control cells.
Figure 6
CHD4 controls the G1/S cell-cycle transition through p53 deacetylation. (A) Fluorescence-activated cell sorting (FACS) profiles of U2OS cells downregulated for CHD4 (siCHD4) compared with control (siLuci). Percentages of cells in G1 are indicated. Nocodazole was used to block cell-cycle progression in mitosis (bottom panels). (B) Western-blot analysis of total extracts from U2OS cells downregulated for CHD4 (siCHD4) compared with control (siLuci) using the indicated antibodies. (C) Quantitative RT–PCR analysis of p21, p53 and β-actin mRNA levels in U2OS cells downregulated for CHD4 (siCHD4) compared with control (siLuci). Error bars indicate s.d. from two independent experiments. (D) Western-blot analysis of p21 induction on total extracts from p53 proficient or deficient HCT116 cells upon CHD4 depletion (C) compared with control siLuciferase (L). + IR: 9 h post-exposure to 10 Gy IR. (E, F) Western-blot analysis of total extracts from U2OS cells treated with the indicated siRNAs. Phleo: 1 h post-exposure to 60 μg/ml Phleomycin. (G) FACS profiles of U2OS cells treated with the indicated siRNAs. Nocodazole was used to block cell-cycle progression in mitosis.
Similar articles
- Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment.
Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. Pan MR, et al. J Biol Chem. 2012 Feb 24;287(9):6764-72. doi: 10.1074/jbc.M111.287037. Epub 2012 Jan 4. J Biol Chem. 2012. PMID: 22219182 Free PMC article. - CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now.
O'Shaughnessy A, Hendrich B. O'Shaughnessy A, et al. Biochem Soc Trans. 2013 Jun;41(3):777-82. doi: 10.1042/BST20130027. Biochem Soc Trans. 2013. PMID: 23697937 Free PMC article. Review. - The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
Silva AP, Ryan DP, Galanty Y, Low JK, Vandevenne M, Jackson SP, Mackay JP. Silva AP, et al. J Biol Chem. 2016 Jan 8;291(2):924-38. doi: 10.1074/jbc.M115.683227. Epub 2015 Nov 12. J Biol Chem. 2016. PMID: 26565020 Free PMC article. - CHD4 Is a Peripheral Component of the Nucleosome Remodeling and Deacetylase Complex.
Low JK, Webb SR, Silva AP, Saathoff H, Ryan DP, Torrado M, Brofelth M, Parker BL, Shepherd NE, Mackay JP. Low JK, et al. J Biol Chem. 2016 Jul 22;291(30):15853-66. doi: 10.1074/jbc.M115.707018. Epub 2016 May 27. J Biol Chem. 2016. PMID: 27235397 Free PMC article. - An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.
Banerjee S, Roy S. Banerjee S, et al. Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
- SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability.
Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, Liang J, Cheng Z, Shi L, Shang Y, Yu W. Li L, et al. Nat Commun. 2016 Jul 20;7:12235. doi: 10.1038/ncomms12235. Nat Commun. 2016. PMID: 27436229 Free PMC article. - The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair.
Roos WP, Krumm A. Roos WP, et al. Nucleic Acids Res. 2016 Dec 1;44(21):10017-10030. doi: 10.1093/nar/gkw922. Epub 2016 Oct 13. Nucleic Acids Res. 2016. PMID: 27738139 Free PMC article. Review. - The Tale of CHD4 in DNA Damage Response and Chemotherapeutic Response.
Zhang J, Shih DJH, Lin SY. Zhang J, et al. J Cancer Res Cell Ther. 2019;3(1):052. Epub 2019 Jul 8. J Cancer Res Cell Ther. 2019. PMID: 32577620 Free PMC article. - ATP-dependent chromatin remodeling in the DNA-damage response.
Lans H, Marteijn JA, Vermeulen W. Lans H, et al. Epigenetics Chromatin. 2012 Jan 30;5:4. doi: 10.1186/1756-8935-5-4. Epigenetics Chromatin. 2012. PMID: 22289628 Free PMC article. - Small molecule inhibitors targeting the cancers.
Liu GH, Chen T, Zhang X, Ma XL, Shi HS. Liu GH, et al. MedComm (2020). 2022 Oct 13;3(4):e181. doi: 10.1002/mco2.181. eCollection 2022 Dec. MedComm (2020). 2022. PMID: 36254250 Free PMC article. Review.
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC (2008) Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 - PubMed
- Bao Y, Shen X (2007) Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev 17: 126–131 - PubMed
- Caldecott KW (2008) Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 - PubMed
- Carter S, Vousden KH (2009) Modifications of p53: competing for the lysines. Curr Opin Genet Dev 19: 18–24 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- A5290/CRUK_/Cancer Research UK/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- A4361/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 11224/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous