The cell biology of taste - PubMed (original) (raw)
Review
The cell biology of taste
Nirupa Chaudhari et al. J Cell Biol. 2010.
Erratum in
- J Cell Biol. 2010 Oct 18;191(2):429
Abstract
Taste buds are aggregates of 50-100 polarized neuroepithelial cells that detect nutrients and other compounds. Combined analyses of gene expression and cellular function reveal an elegant cellular organization within the taste bud. This review discusses the functional classes of taste cells, their cell biology, and current thinking on how taste information is transmitted to the brain.
Figures
Figure 1.
Taste qualities, the taste receptors that detect them, and examples of natural stimuli. Five recognized taste qualities—sweet, sour, bitter, salty, and umami—are detected by taste buds. Bitter taste is thought to protect against ingesting poisons, many of which taste bitter. Sweet taste signals sugars and carbohydrates. Umami taste is elicited by
l
-amino acids and nucleotides. Salty taste is generated mainly by Na+ and sour taste potently by organic acids. Evidence is mounting that fat may also be detected by taste buds via dedicated receptors. The names of taste receptors and cartoons depicting their transmembrane topology are shown outside the perimeter. Bitter is transduced by G protein–coupled receptors similar to Class I GPCRs (with short extracellular N termini). In contrast, sweet and umami are detected by dimers of Class III GPCRs (with long N termini that form a globular extracellular ligand-binding domain). One of the receptors for Na+ salts is a cation channel composed of three subunits, each with two transmembrane domains. Membrane receptors for sour and fat are as yet uncertain.
Figure 2.
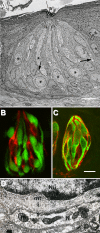
Cell types and synapses in the taste bud. (A) Electron micrograph of a rabbit taste bud showing cells with dark or light cytoplasm, and nerve profiles (arrows). Asterisks mark Type II (receptor) cells. Reprinted with permission from J. Comp. Neurol. (Royer and Kinnamon, 1991). (B) A taste bud from a transgenic mouse expressing GFP only in receptor (Type II) cells. Presynaptic cells are immunostained (red) for aromatic amino acid decarboxylase (a neurotransmitter-synthesizing enzyme that is a marker for these cells), and are distinct from receptor cells, identified by GFP (green). Reprinted with permission from J. Neurosci. (C) Taste buds immunostained for NTPDase2 (an ectonucleotidase associated with the plasma membrane of Type I cells) reveal the thin lamellae (red) of Type I cells. These cytoplasmic extensions wrap around other cells in the taste bud. GFP (green) indicates receptor cells as in B. Bar, 10 µm. Image courtesy of M.S. Sinclair and N. Chaudhari. (D) High magnification electron micrograph of a synapse between a presynaptic taste cell and a nerve terminal (N) in a hamster taste bud. The nucleus (Nu) of the presynaptic cell is at the top, and neurotransmitter vesicles cluster near the synapse(s). The nerve profile includes mitochondria (m) and electron-dense postsynaptic densities. mt, microtubule. Image courtesy of J.C. Kinnamon.
Figure 3.
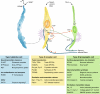
The three major classes of taste cells. This classification incorporates ultrastructural features, patterns of gene expression, and the functions of each of Types I, II (receptor), and III (presynaptic) taste cells. Type I cells (blue) degrade or absorb neurotransmitters. They also may clear extracellular K+ that accumulates after action potentials (shown as bursts) in receptor (yellow) and presynaptic (green) cells. K+ may be extruded through an apical K channel such as ROMK. Salty taste may be transduced by some Type I cells, but this remains uncertain. Sweet, bitter, and umami taste compounds activate receptor cells, inducing them to release ATP through pannexin1 (Panx1) hemichannels. The extracellular ATP excites ATP receptors (P2X, P2Y) on sensory nerve fibers and on taste cells. Presynaptic cells, in turn, release serotonin (5-HT), which inhibits receptor cells. Sour stimuli (and carbonation, not depicted) directly activate presynaptic cells. Only presynaptic cells form ultrastructurally identifiable synapses with nerves. Tables below the cells list some of the proteins that are expressed in a cell type–selective manner.
Figure 4.
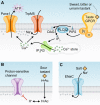
Mechanisms by which five taste qualities are transduced in taste cells. (A) In receptor (Type II) cells, sweet, bitter, and umami ligands bind taste GPCRs, and activate a phosphoinositide pathway that elevates cytoplasmic Ca2+ and depolarizes the membrane via a cation channel, TrpM5. The combined action of elevated Ca2+ and membrane depolarization opens the large pores of gap junction hemichannels, likely composed of Panx1, resulting in ATP release. Shown here is a dimer of T1R taste GPCRs (sweet, umami). T2R taste GPCRs (bitter) do not have extensive extracellular domains and it is not known whether T2Rs form multimers. (B) In presynaptic (Type III) cells, organic acids (HAc) permeate through the plasma membrane and acidify the cytoplasm where they dissociate to acidify the cytosol. Intracellular H+ is believed to block a proton-sensitive K channel (as yet unidentified) and depolarize the membrane. Voltage-gated Ca channels would then elevate cytoplasmic Ca2+ to trigger exocytosis of synaptic vesicles (not depicted). (C) The salty taste of Na+ is detected by direct permeation of Na+ ions through membrane ion channels, including ENaC, to depolarize the membrane. The cell type underlying salty taste has not been definitively identified.
Similar articles
- The cell biology of vertebrate taste receptors.
Roper SD. Roper SD. Annu Rev Neurosci. 1989;12:329-53. doi: 10.1146/annurev.ne.12.030189.001553. Annu Rev Neurosci. 1989. PMID: 2648951 Review. - Processing umami and other tastes in mammalian taste buds.
Roper SD, Chaudhari N. Roper SD, et al. Ann N Y Acad Sci. 2009 Jul;1170:60-5. doi: 10.1111/j.1749-6632.2009.04107.x. Ann N Y Acad Sci. 2009. PMID: 19686109 Free PMC article. Review. - Human taste: response and taste bud number in fungiform papillae.
Arvidson K, Friberg U. Arvidson K, et al. Science. 1980 Aug 15;209(4458):807-8. doi: 10.1126/science.7403846. Science. 1980. PMID: 7403846 - The gustatory system of lampreys.
Barreiro-Iglesias A, Anadón R, Rodicio MC. Barreiro-Iglesias A, et al. Brain Behav Evol. 2010;75(4):241-50. doi: 10.1159/000315151. Epub 2010 Jul 24. Brain Behav Evol. 2010. PMID: 20664239 Review. - Electrophysiology of Necturus taste cells.
Bigiani A. Bigiani A. Prog Neurobiol. 2002 Feb;66(3):123-59. doi: 10.1016/s0301-0082(02)00005-9. Prog Neurobiol. 2002. PMID: 11943449 Review.
Cited by
- Recent Advances in Artificial Sensory Neurons: Biological Fundamentals, Devices, Applications, and Challenges.
Zhong S, Su L, Xu M, Loke D, Yu B, Zhang Y, Zhao R. Zhong S, et al. Nanomicro Lett. 2024 Nov 13;17(1):61. doi: 10.1007/s40820-024-01550-x. Nanomicro Lett. 2024. PMID: 39537845 Free PMC article. Review. - Bitter taste receptors as sensors of gut luminal contents.
Sternini C, Rozengurt E. Sternini C, et al. Nat Rev Gastroenterol Hepatol. 2024 Oct 28. doi: 10.1038/s41575-024-01005-z. Online ahead of print. Nat Rev Gastroenterol Hepatol. 2024. PMID: 39468215 Review. - The Expression of Cannabinoid and Cannabinoid-Related Receptors on the Gustatory Cells of the Piglet Tongue.
Zamith Cunha R, Grilli E, Piva A, Delprete C, Franciosi C, Caprini M, Chiocchetti R. Zamith Cunha R, et al. Molecules. 2024 Sep 28;29(19):4613. doi: 10.3390/molecules29194613. Molecules. 2024. PMID: 39407543 Free PMC article. - Death in the taste bud: Morphological features of dying taste cells and engulfment by Type I cells.
Wilson CE, Lasher RS, Salcedo E, Yang R, Dzowo Y, Kinnamon JC, Finger TE. Wilson CE, et al. bioRxiv [Preprint]. 2024 Sep 11:2024.09.06.611711. doi: 10.1101/2024.09.06.611711. bioRxiv. 2024. PMID: 39314340 Free PMC article. Preprint. - Bitter tastants relax the mouse gallbladder smooth muscle independent of signaling through tuft cells and bitter taste receptors.
Keshavarz M, Ruppert AL, Meiners M, Poharkar K, Liu S, Mahmoud W, Winterberg S, Hartmann P, Mermer P, Perniss A, Offermanns S, Kummer W, Schütz B. Keshavarz M, et al. Sci Rep. 2024 Aug 8;14(1):18447. doi: 10.1038/s41598-024-69287-6. Sci Rep. 2024. PMID: 39117690 Free PMC article.
References
- Behrens M., Foerster S., Staehler F., Raguse J.D., Meyerhof W. 2007. Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogenous population of bitter responsive taste receptor cells. J. Neurosci. 27:12630–12640 10.1523/JNEUROSCI.1168-07.2007 - DOI - PMC - PubMed
- Bigiani A. 2001. Mouse taste cells with glialike membrane properties. J. Neurophysiol. 85:1552–1560 - PubMed
Publication types
MeSH terms
Grants and funding
- R01 DC006308/DC/NIDCD NIH HHS/United States
- R56 DC006308/DC/NIDCD NIH HHS/United States
- R01 DC006021/DC/NIDCD NIH HHS/United States
- DC010073/DC/NIDCD NIH HHS/United States
- R55 DC007630/DC/NIDCD NIH HHS/United States
- R21 DC010073/DC/NIDCD NIH HHS/United States
- R01 DC007630/DC/NIDCD NIH HHS/United States
- DC006308/DC/NIDCD NIH HHS/United States
- DC006021/DC/NIDCD NIH HHS/United States
- F32 DC000374/DC/NIDCD NIH HHS/United States
- R01 DC000374/DC/NIDCD NIH HHS/United States
- R01 DC006308-06A1/DC/NIDCD NIH HHS/United States
- DC007630/DC/NIDCD NIH HHS/United States
- R21 DC010073-02/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources