Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat - PubMed (original) (raw)
Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat
James R R Whittle et al. J Cell Biol. 2010.
Abstract
Ancestral coatomer element 1 (ACE1) proteins assemble latticework coats for COPII vesicles and the nuclear pore complex. The ACE1 protein Sec31 and Sec13 make a 2:2 tetramer that forms the edge element of the COPII outer coat. In this study, we report that the COPII accessory protein Sec16 also contains an ACE1. The 165-kD crystal structure of the central domain of Sec16 in complex with Sec13 was solved at 2.7-A resolution. Sec16 and Sec13 also make a 2:2 tetramer, another edge element for the COPII system. Domain swapping at the ACE1-ACE1 interface is observed both in the prior structure of Sec13-Sec31 and in Sec13-Sec16. A Sec31 mutant in which domain swapping is prevented adopts an unprecedented laminated structure, solved at 2.8-A resolution. Our in vivo data suggest that the ACE1 element of Sec31 can functionally replace the ACE1 element of Sec16. Our data support Sec16 as a scaffold for the COPII system and a template for the Sec13-Sec31 coat.
Figures
Figure 1.
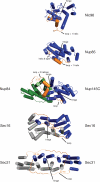
Summary of ancestral coatomer element (ACE1) proteins. ACE1 was originally identified based on structural homology between Nup85, Nic96, Nup84, Nup145C, and Sec31. Sec16 is shown to contain an ACE1 in this study. Three ACE1 proteins bind Sec13, mutually exclusively; Nup85 binds its homologue Seh1. Nup84 and Nup145C form a heterodimer, and Sec16 and Sec31 form homodimers. The structure of Nic96 is shown to illustrate the three modules that compose the ACE1: crown, trunk, and tail. The COPII ACE1 domains might lack the tail module. The structure is colored red to white from the N to C terminus, as labeled. Dashed arrows show how the ACE1 forms a J shape. A dotted arc encircles the surface by which ACE1 dimerization occurs.
Figure 2.
Structural comparison of Sec16 and Sec31. (A) Domain organization of Sec16. Unstructured regions (wavy black lines), CCD (blue–cyan box), and C-terminal α-helical domain (white box, labeled αH) are shown. Scale bar shows number of amino acids residues. The β-propeller protein Sec13 binds Sec16 at the position indicated. (B) Crystal structure of Sec16 in complex with Sec13. Sec13 is colored orange to yellow from the N to C terminus, as in A. One Sec16 monomer is colored blue to cyan, as in A, the other dark to light gray from the N to C terminus. N and C termini are labeled, as well as β-strands β1–β3 and helices α0–α18. Helices α5–α7 of the Sec16 monomer domain swap at the homotypic interface. The bottom view is rotated 90° toward the viewer as indicated. (C) Domain organization of Sec31. Unstructured region (wavy black line), β-propeller (white oval, labeled βPROP), central structured domain (blue–cyan box), and C-terminal α-helical domain (white box, labeled αH) are shown. Scale bar shows number of amino acids residues. The β-propeller protein Sec13 binds Sec31 at the position indicated. (D) Structure of the Sec13–Sec31 tetramer. Color scheme as in B, with Sec31 coloring matching that of Sec16. The bottom view is rotated 90° toward the viewer as indicated.
Figure 3.
Interactions formed by the ACE1 crown. The crown domain of each ACE1 is shown colored blue, except Nup84, which is green. Nic96 and Nup85 are not known to dimerize via crown–crown interaction. Nup84 and Nup145C form a heterodimer, and Sec16 and Sec31 form homodimers. The second copies of Sec16 and Sec31 are colored gray. Disordered loops are shown as dotted lines and labeled with the number of amino acids not observed. Sec16 and Sec31 dimerize by domain swapping. Helices α5–α7 exchange positions with helices α5′–α7′ in the binding partner. The domain swap requires extension of the swap loop (labeled loop) that connects helix α4 to α5 and rotation of the swap hinge (labeled hinge) that connects helix α7 to α8. The positions of the corresponding loop and hinge are labeled in all ACE1s. Nic96, Nup85, and Nup84 have one or two α-helices inserted into this loop (labeled loop + n helices).
Figure 4.
Solution behavior and crystal structure of Sec13–Sec31 mutants. (A) Sec13–Sec31 (red) compared with Sec13–Sec31 mutants ΔL (blue) and EE (green) by size exclusion chromatography. Sec13–Sec31 and Sec13–Sec31ΔL are heterotetramers, whose structures are shown in B, with the same hydrodynamic radii. The EE mutation disrupts the crown interface. Sec13–Sec31EE is a heterodimer in solution. Elution volume in milliliters is plotted against absorbance at λ = 280 nm. Elution volumes for standard globular proteins are as indicated. SDS-PAGE analysis of each sample is shown, with fractions indicated below the x axis. (B) Crystal structure of Sec13–Sec31ΔL compared with Sec13–Sec31 (Protein Data Bank ID 2PM6). The tetramers form extended rods with the same 165° central angle. Because of deletion of the swap loop, Sec13–Sec31ΔL forms a laminated structure, rather than a U turn, as indicated by arrows labeled N and C. Both copies of Sec31 are colored blue to cyan from the N to C terminus. The green label EE indicates residues M540 and L544, in helices α7 and α7′, which were mutated to glutamic acid to generate Sec13–Sec31EE.
Figure 5.
Complementation of Sec16 or Sec31 by structure-based mutations. (A) Structure-based mutants of Sec31 assayed by plasmid shuffle. A sec31Δ plasmid shuffle strain was prepared using endogenous SEC31 cloned into a URA3 plasmid. Mutations were introduced into SEC31 on a LEU2 plasmid, transformed into the shuffle strain, spotted onto media lacking leucine (first lane) or lacking leucine and supplemented with 5-fluoroorotic acid (5-FOA; subsequent lanes), and grown for 36 h at 24, 30, or 37°C. Sec31 domain architecture is diagramed: N-terminal β-propeller (white oval), central insertion β-blade and ACE1 (red–white box), unstructured region (wavy black lines), and C-terminal α-helical domain (white box). Fragments of Sec16 used in chimeric genes are shown in blue–cyan. (B) Structure-based mutants of Sec16 assayed by plasmid shuffle. Mutations were introduced into SEC16 on a LEU2 plasmid and tested as in A. Genes are diagramed: unstructured regions (wavy black lines), CCD (blue–cyan box), and C-terminal α-helical domain (white box). Fragments of Sec31 used in chimeric genes are shown in red–white.
Figure 6.
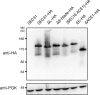
Western blot of structure-based mutants of Sec31. Structure-based mutants of Sec31 were tagged with the HA epitope and transformed into the sec31Δ plasmid shuffle strain (as in Fig. 5 A). Untagged Sec31 is shown as a control for specificity of the anti-HA antibody. The blot was reprobed with an antibody to 3-phosphoglycerate kinase (PGK) as a control for equal sample loading. MM, molecular mass.
Figure 7.
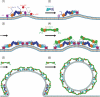
Model for assembly of the COPII coat complex. The common model for assembly of the COPII coat complex is modified to include the role of Sec16. For simplicity, cargo molecules are omitted. (1) The Sec13–Sec16 tetramer is stably associated with the ER membrane and binds the integral membrane protein Sed4 or its homologue Sec12. Sar1 becomes associated with the membrane, when it is converted from the GDP- to GTP-bound state. Concentration of membrane-associated proteins begins to bend membrane. (2) Sec13–Sec16 and Sar1 collaborate to recruit the cargo adaptor Sec23–Sec24 dimer. (3) A precoat self-associates into higher-order oligomers. (4) Sec13–Sec16 and Sec23–Sec24–Sar1 form independent interactions with Sec13–Sec31, causing it to assemble near and/or in place of Sec16. (5) The forming coat contains progressively more Sec13–Sec31 and less Sec13–Sec16. Hand-off of Sec23–Sec24–Sar1 from Sec16 to Sec31 sets the stage for GTP hydrolysis by Sar1. (6) A final COPII coat is formed, and vesicle budding is complete. Sec13–Sec16 remains mostly associated with the ER.
Similar articles
- ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p.
Copic A, Latham CF, Horlbeck MA, D'Arcangelo JG, Miller EA. Copic A, et al. Science. 2012 Mar 16;335(6074):1359-62. doi: 10.1126/science.1215909. Epub 2012 Feb 2. Science. 2012. PMID: 22300850 Free PMC article. - Making COPII coats.
Kirchhausen T. Kirchhausen T. Cell. 2007 Jun 29;129(7):1251-2. doi: 10.1016/j.cell.2007.06.015. Cell. 2007. PMID: 17604713 Review. - Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - Structure and organization of coat proteins in the COPII cage.
Fath S, Mancias JD, Bi X, Goldberg J. Fath S, et al. Cell. 2007 Jun 29;129(7):1325-36. doi: 10.1016/j.cell.2007.05.036. Cell. 2007. PMID: 17604721 - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review.
Cited by
- Vesicle-mediated export from the ER: COPII coat function and regulation.
D'Arcangelo JG, Stahmer KR, Miller EA. D'Arcangelo JG, et al. Biochim Biophys Acta. 2013 Nov;1833(11):2464-72. doi: 10.1016/j.bbamcr.2013.02.003. Epub 2013 Feb 15. Biochim Biophys Acta. 2013. PMID: 23419775 Free PMC article. Review. - Loss of hepatic VMP1 trapped VLDL in the bilayer of endoplasmic reticulum membrane☆.
Ni HM, Ding B, Chen A. Ni HM, et al. Liver Res. 2023 Jun;7(2):161-163. doi: 10.1016/j.livres.2023.04.001. Epub 2023 May 3. Liver Res. 2023. PMID: 38405163 Free PMC article. No abstract available. - A pseudoatomic model of the COPII cage obtained from cryo-electron microscopy and mass spectrometry.
Noble AJ, Zhang Q, O'Donnell J, Hariri H, Bhattacharya N, Marshall AG, Stagg SM. Noble AJ, et al. Nat Struct Mol Biol. 2013 Feb;20(2):167-73. doi: 10.1038/nsmb.2467. Epub 2012 Dec 23. Nat Struct Mol Biol. 2013. PMID: 23262493 Free PMC article. - Mechanisms for exporting large-sized cargoes from the endoplasmic reticulum.
Saito K, Katada T. Saito K, et al. Cell Mol Life Sci. 2015 Oct;72(19):3709-20. doi: 10.1007/s00018-015-1952-9. Epub 2015 Jun 17. Cell Mol Life Sci. 2015. PMID: 26082182 Free PMC article. Review. - TFG regulates inner COPII coat recruitment to facilitate anterograde secretory protein transport.
Kasberg W, Luong P, Minushkin K, Pustova I, Swift KA, Zhao M, Audhya A. Kasberg W, et al. Mol Biol Cell. 2024 Aug 1;35(8):ar113. doi: 10.1091/mbc.E24-06-0282. Epub 2024 Jul 10. Mol Biol Cell. 2024. PMID: 38985515 Free PMC article.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 10.1107/S0907444902016657 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases