Describing the mechanism of antimicrobial peptide action with the interfacial activity model - PubMed (original) (raw)
Review
Describing the mechanism of antimicrobial peptide action with the interfacial activity model
William C Wimley. ACS Chem Biol. 2010.
Abstract
Antimicrobial peptides (AMPs) have been studied for three decades, and yet a molecular understanding of their mechanism of action is still lacking. Here we summarize current knowledge for both synthetic vesicle experiments and microbe experiments, with a focus on comparisons between the two. Microbial experiments are done at peptide to lipid ratios that are at least 4 orders of magnitude higher than vesicle-based experiments. To close the gap between the two concentration regimes, we propose an "interfacial activity model", which is based on an experimentally testable molecular image of AMP-membrane interactions. The interfacial activity model may be useful in driving engineering and design of novel AMPs.
Figures
Figure 1
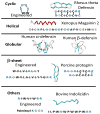
A menagerie of antimicrobial peptide structures. AMPs range from 4 to about 40 amino acids in length, Some are linear, while others are cyclic, disulfide crosslinked or acylated. Known antimicrobial peptides have many types of secondary structure, including α-helix, β-sheet and irregular structure or random coil. Engineered AMPs have the same properties as natural examples. The one feature that unites all AMPs is their hydrophobic and cationic nature.
Figure 2
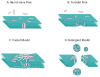
Commonly cited models for antimicrobial peptide activity. Barrel-stave and toroidal pores are membrane-spanning aqueous channels. Antimicrobial peptides are described with the carpet model. Such peptides permeabilize membranes by “carpeting” the bilayer with peptides. At high concentrations carpet model peptides can behave more like detergents.
Figure 3
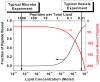
Peptide activity against lipid vesicles. True transmembrane pore-forming peptides, such as alamethicin, permeabilize vesicles at very low peptide:lipid ratios. The green line is based on experimental measurements. Antimicrobial peptides, on the other hand, are active against lipid vesicles only at high peptide:lipid ratios. Data points in red are actual data from several recent publications(13;17). Almost any peptide that binds to membranes can cause leakage at very high concentration, shown schematically in blue.
Figure 4
Release of vesicle-entrapped probes. A: A large unilamellar vesicle of 0.1 μm LUV, the type used in most experiments, is made of roughly 105 lipids enclosing a volume of about 10−19 liter. B: Predicted kinetics of probe release from large unilamellar vesicles containing a single pore of 10 Å diameter. Complete release occurs in a few tenths of a second. C: Actual release kinetics observed in experiments with AMPs. Note the difference in X-axis scale indicating that actual release data is 3 orders of magnitude slower than predicted. Furthermore, the simulated release goes to completion, while the actual release is incomplete.
Figure 5
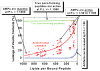
Typical peptide:lipid ratios in vesicle leakage experiments and microbe sterilization assays. Microbe sterilization assays are done under conditions of peptide excess over lipid such that the membrane is saturated by peptide and there is a large reservoir of unbound peptide. Vesicle leakage experiments are done under conditions where lipid is in excess and most peptide is bound.
Figure 6
Molecular dynamics simulation of peptide pore formation from Sengupta and Marrink (65). Top: The peptide-lipid bilayer with CH2 groups removed. Blue spheres are water molecules and yellow spheres are the terminal methyl groups. Other color spheres are the lipid polar groups. Bottom: The same peptide-lipid bilayer separated into groups. Notice in the vicinity of the peptide “pore” the strict segregation between polar and non-polar is broken down.
Similar articles
- Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity.
Rathinakumar R, Walkenhorst WF, Wimley WC. Rathinakumar R, et al. J Am Chem Soc. 2009 Jun 10;131(22):7609-17. doi: 10.1021/ja8093247. J Am Chem Soc. 2009. PMID: 19445503 Free PMC article. - (Lipo)polysaccharide interactions of antimicrobial peptides.
Schmidtchen A, Malmsten M. Schmidtchen A, et al. J Colloid Interface Sci. 2015 Jul 1;449:136-42. doi: 10.1016/j.jcis.2014.11.024. Epub 2014 Nov 20. J Colloid Interface Sci. 2015. PMID: 25490856 Review. - Alpha-helical cationic antimicrobial peptides: relationships of structure and function.
Huang Y, Huang J, Chen Y. Huang Y, et al. Protein Cell. 2010 Feb;1(2):143-52. doi: 10.1007/s13238-010-0004-3. Epub 2010 Feb 6. Protein Cell. 2010. PMID: 21203984 Free PMC article. Review. - A Review of the Mechanism of Action of Amphibian Antimicrobial Peptides Focusing on Peptide-Membrane Interaction and Membrane Curvature.
Vineeth Kumar TV, Sanil G. Vineeth Kumar TV, et al. Curr Protein Pept Sci. 2017;18(12):1263-1272. doi: 10.2174/1389203718666170710114932. Curr Protein Pept Sci. 2017. PMID: 28699512 Review. - Effect of stereochemistry, chain length and sequence pattern on antimicrobial properties of short synthetic β-sheet forming peptide amphiphiles.
Ong ZY, Cheng J, Huang Y, Xu K, Ji Z, Fan W, Yang YY. Ong ZY, et al. Biomaterials. 2014 Jan;35(4):1315-25. doi: 10.1016/j.biomaterials.2013.10.053. Epub 2013 Nov 7. Biomaterials. 2014. PMID: 24211081
Cited by
- Plant antimicrobial peptides: structures, functions, and applications.
Li J, Hu S, Jian W, Xie C, Yang X. Li J, et al. Bot Stud. 2021 Apr 29;62(1):5. doi: 10.1186/s40529-021-00312-x. Bot Stud. 2021. PMID: 33914180 Free PMC article. Review. - The multifaceted nature of antimicrobial peptides: current synthetic chemistry approaches and future directions.
Gan BH, Gaynord J, Rowe SM, Deingruber T, Spring DR. Gan BH, et al. Chem Soc Rev. 2021 Jul 5;50(13):7820-7880. doi: 10.1039/d0cs00729c. Chem Soc Rev. 2021. PMID: 34042120 Free PMC article. Review. - Melittin can permeabilize membranes via large transient pores.
Ulmschneider JP, Ulmschneider MB. Ulmschneider JP, et al. Nat Commun. 2024 Aug 23;15(1):7281. doi: 10.1038/s41467-024-51691-1. Nat Commun. 2024. PMID: 39179607 Free PMC article. - A membrane-translocating peptide penetrates into bilayers without significant bilayer perturbations.
Cruz J, Mihailescu M, Wiedman G, Herman K, Searson PC, Wimley WC, Hristova K. Cruz J, et al. Biophys J. 2013 Jun 4;104(11):2419-28. doi: 10.1016/j.bpj.2013.04.043. Biophys J. 2013. PMID: 23746514 Free PMC article. - pH Dependence of microbe sterilization by cationic antimicrobial peptides.
Walkenhorst WF, Klein JW, Vo P, Wimley WC. Walkenhorst WF, et al. Antimicrob Agents Chemother. 2013 Jul;57(7):3312-20. doi: 10.1128/AAC.00063-13. Epub 2013 May 6. Antimicrob Agents Chemother. 2013. PMID: 23650166 Free PMC article.
References
- Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature (London) 1981;292:246–248. - PubMed
- Tossi A. Antimicrobial peptides datatbase. 2005. http://www.bbcm.units.it/~tossi/. Web Site.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous