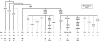Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean - PubMed (original) (raw)
Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean
Said Msaidie et al. Eur J Hum Genet. 2011 Jan.
Abstract
The Comoros Islands are situated off the coast of East Africa, at the northern entrance of the channel of Mozambique. Contemporary Comoros society displays linguistic, cultural and religious features that are indicators of interactions between African, Middle Eastern and Southeast Asian (SEA) populations. Influences came from the north, brought by the Arab and Persian traders whose maritime routes extended to Madagascar by 700-900 AD. Influences also came from the Far East, with the long-distance colonisation by Austronesian seafarers that reached Madagascar 1500 years ago. Indeed, strong genetic evidence for a SEA, but not a Middle Eastern, contribution has been found on Madagascar, but no genetic trace of either migration has been shown to exist in mainland Africa. Studying genetic diversity on the Comoros Islands could therefore provide new insights into human movement in the Indian Ocean. Here, we describe Y chromosomal and mitochondrial genetic variation in 577 Comorian islanders. We have defined 28 Y chromosomal and 9 mitochondrial lineages. We show the Comoros population to be a genetic mosaic, the result of tripartite gene flow from Africa, the Middle East and Southeast Asia. A distinctive profile of African haplogroups, shared with Madagascar, may be characteristic of coastal sub-Saharan East Africa. Finally, the absence of any maternal contribution from Western Eurasia strongly implicates male-dominated trade and religion as the drivers of gene flow from the North. The Comoros provides a first view of the genetic makeup of coastal East Africa.
Figures
Figure 1
Frequencies (%) and numbers (n) of Y haplogroups in the Comoros population sample. Haplogroup names follow the 2008 nomenclature. Branches are labelled with the binary markers tested. Numbers without a letter represent ‘M' prefixed Y markers (eg, 50=M50). Putative geographic origin is indicated for each haplogroup: Af – sub-Saharan Africa, WSA – West and Southwest Asia, SEA – Southeast Asia and ? – uncertain. Frequencies of less than 5% have been rounded up or down to the nearest unit.
Figure 2
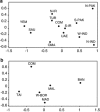
Multidimensional scaling (MDS) analysis plot of genetic distance (Rst) calculated from the incidence of alleles at eight Y-STR loci (DYS19, 389AB, 389CD, 390, 391, 392, 393, 439). The analysis was performed with subsets of the Comoros sample, which were created on the basis of putative haplogroup origin. (a) Middle East – haplogroups E-M123, E-V22, F, G, J, L, Q and R. (b) Southeast Asian – haplogroups O, C* and K*. The populations represented are the Comoros (COM), this study, Madagascar (MAD), Oman (OMA),, Turkey (TUR), North Pakistan (N-PAK), South Pakistan (S-PAK), North India (N-IND), South India (S-IND), Yemen (YEM), United Arab Emirates (UAE), Saudi Arabia (SAU), North Iran (N-IR), South Iran (S-IR),, Malaysia (MAL), Taiwan (Paiwan) (TAI), West Borneo (East Malaysia) (W-BOR) and Bangladesh (BAN).
Figure 3
Frequencies (%) and numbers (n) of mitochondrial haplogroups in the Comoros population sample. Numbers on branches refer to the position of polymorphisms in the CRS (Cambridge reference sequence). HVS-I sequence was not determined for L0, L1, L2 or L3′4(xMN). The HVS-I SNPs are shown for M and N haplogroups, only where they provide further definition than the coding SNPs. Putative geographic origin is indicated for each haplogroup: Af – sub-Saharan Africa, SEA – Southeast Asia and ? – uncertain.
Figure 4
Multidimensional scaling (MDS) analysis plot of genetic distance (Fst) calculated from mitochondrial haplogroup frequencies. M* and R* were excluded from these analyses. (a) Africa, SEA and Iran – all Comoros haplogroups, except M* and R*. (b) and (c) MDS performed with subsets of the Comoros sample, defined on the basis of putative haplogroup origin. (b) Africa – Comoros haplogroups L. (c) SEA – Comoros and Madagascar haplogroups B4a, B4a1a1-PM, F3b, M7c1c and R9. The populations are Comoros (COM), this study, Madagascar (MAD), Central Africa (AFC), Iran (IRA), Mozambique (MOZ), Kenya (KEN), Ethopia (ETH), Tunisia (TUN), Algeria (ALG), Morocco (MOR), Mauritania (MAU), Taiwan (TAI), Philippines (PHI), Malaysia (MAL), Borneo (BOR), Sumatra (SUM), Bali (BAL) and Java (JAV).
Similar articles
- Evidence of Austronesian Genetic Lineages in East Africa and South Arabia: Complex Dispersal from Madagascar and Southeast Asia.
Brucato N, Fernandes V, Kusuma P, Černý V, Mulligan CJ, Soares P, Rito T, Besse C, Boland A, Deleuze JF, Cox MP, Sudoyo H, Stoneking M, Pereira L, Ricaut FX. Brucato N, et al. Genome Biol Evol. 2019 Mar 1;11(3):748-758. doi: 10.1093/gbe/evz028. Genome Biol Evol. 2019. PMID: 30715341 Free PMC article. - The Comoros Show the Earliest Austronesian Gene Flow into the Swahili Corridor.
Brucato N, Fernandes V, Mazières S, Kusuma P, Cox MP, Ng'ang'a JW, Omar M, Simeone-Senelle MC, Frassati C, Alshamali F, Fin B, Boland A, Deleuze JF, Stoneking M, Adelaar A, Crowther A, Boivin N, Pereira L, Bailly P, Chiaroni J, Ricaut FX. Brucato N, et al. Am J Hum Genet. 2018 Jan 4;102(1):58-68. doi: 10.1016/j.ajhg.2017.11.011. Am J Hum Genet. 2018. PMID: 29304377 Free PMC article. - The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages.
Hurles ME, Sykes BC, Jobling MA, Forster P. Hurles ME, et al. Am J Hum Genet. 2005 May;76(5):894-901. doi: 10.1086/430051. Epub 2005 Mar 25. Am J Hum Genet. 2005. PMID: 15793703 Free PMC article. - The human genetic history of East Asia: weaving a complex tapestry.
Stoneking M, Delfin F. Stoneking M, et al. Curr Biol. 2010 Feb 23;20(4):R188-93. doi: 10.1016/j.cub.2009.11.052. Curr Biol. 2010. PMID: 20178766 Review. - Ancient migration routes of Austronesian-speaking populations in oceanic Southeast Asia and Melanesia might mimic the spread of nasopharyngeal carcinoma.
Trejaut J, Lee CL, Yen JC, Loo JH, Lin M. Trejaut J, et al. Chin J Cancer. 2011 Feb;30(2):96-105. doi: 10.5732/cjc.010.10589. Chin J Cancer. 2011. PMID: 21272441 Free PMC article. Review.
Cited by
- Ecocultural or Biocultural? Towards Appropriate Terminologies in Biocultural Diversity.
Franco FM. Franco FM. Biology (Basel). 2022 Jan 28;11(2):207. doi: 10.3390/biology11020207. Biology (Basel). 2022. PMID: 35205074 Free PMC article. - Coexistence of honeybees with distinct mitochondrial haplotypes and hybridised nuclear genomes on the Comoros Islands.
Okwaro LA, Muli E, Runo SM, Lattorff HMG. Okwaro LA, et al. Naturwissenschaften. 2021 Apr 19;108(3):17. doi: 10.1007/s00114-021-01729-x. Naturwissenschaften. 2021. PMID: 33871694 - Human OMICs and Computational Biology Research in Africa: Current Challenges and Prospects.
Hamdi Y, Zass L, Othman H, Radouani F, Allali I, Hanachi M, Okeke CJ, Chaouch M, Tendwa MB, Samtal C, Mohamed Sallam R, Alsayed N, Turkson M, Ahmed S, Benkahla A, Romdhane L, Souiai O, Tastan Bishop Ö, Ghedira K, Mohamed Fadlelmola F, Mulder N, Kamal Kassim S. Hamdi Y, et al. OMICS. 2021 Apr;25(4):213-233. doi: 10.1089/omi.2021.0004. Epub 2021 Apr 1. OMICS. 2021. PMID: 33794662 Free PMC article. - How history and geography may explain the distribution in the Comorian archipelago of a novel mutation in DNA repair-deficient xeroderma pigmentosum patients.
Sarasin A, Munier P, Cartault F. Sarasin A, et al. Genet Mol Biol. 2019 Dec 13;43(1 suppl 1):e20190046. doi: 10.1590/1678-4685-GMB-2019-0046. eCollection 2019. Genet Mol Biol. 2019. PMID: 31930276 Free PMC article. - Evidence of Austronesian Genetic Lineages in East Africa and South Arabia: Complex Dispersal from Madagascar and Southeast Asia.
Brucato N, Fernandes V, Kusuma P, Černý V, Mulligan CJ, Soares P, Rito T, Besse C, Boland A, Deleuze JF, Cox MP, Sudoyo H, Stoneking M, Pereira L, Ricaut FX. Brucato N, et al. Genome Biol Evol. 2019 Mar 1;11(3):748-758. doi: 10.1093/gbe/evz028. Genome Biol Evol. 2019. PMID: 30715341 Free PMC article.
References
- Beaujard P. The Indian Ocean in Eurasian and African World-Systems before the Sixteenth Century. J World Hist. 2005;16:411–465.
- Chaudhuri KN. Trade and Civilisation in the Indian Ocean: An Economic History from the Rise of Islam to 1750. Cambridge: Cambridge University Press; 1985.
- Allibert C. Le mot ‘ KOMR' dans l'océan indien. Etudes Océan Indien. 2001;31:13–33.
- Blench R.The ethnographic evidence for long-distance contacts between Oceania and East Africain Reade J (ed).: The Indian Ocean in Antiquity London, New York: Kegan Paul, British Museum; 1994461–470.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources