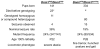A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice - PubMed (original) (raw)
A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice
Rebecca M Boumil et al. PLoS Genet. 2010.
Abstract
Dynamin-1 (Dnm1) encodes a large multimeric GTPase necessary for activity-dependent membrane recycling in neurons, including synaptic vesicle endocytosis. Mice heterozygous for a novel spontaneous Dnm1 mutation--fitful--experience recurrent seizures, and homozygotes have more debilitating, often lethal seizures in addition to severe ataxia and neurosensory deficits. Fitful is a missense mutation in an exon that defines the DNM1a isoform, leaving intact the alternatively spliced exon that encodes DNM1b. The expression of the corresponding alternate transcripts is developmentally regulated, with DNM1b expression highest during early neuronal development and DNM1a expression increasing postnatally with synaptic maturation. Mutant DNM1a does not efficiently self-assemble into higher order complexes known to be necessary for proper dynamin function, and it also interferes with endocytic recycling in cell culture. In mice, the mutation results in defective synaptic transmission characterized by a slower recovery from depression after trains of stimulation. The DNM1a and DNM1b isoform pair is highly conserved in vertebrate evolution, whereas invertebrates have only one isoform. We speculate that the emergence of more specialized forms of DNM1 may be important in organisms with complex neuronal function.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
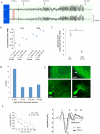
Figure 1. Fitful mice have seizures and neurosensory defects.
(A) Differential traces from EEG recording of fitful heterozygotes. Behavioral events associated with the EEG are shown. Vertical lines are 1s time markings; each channel is approximately 500uV high. (B) Average electroconvulsive thresholds to the minimal forebrain clonic seizure endpoint of B6-Ftfl/+ and B6-+/+ mice at two different stimulus durations (1.0s – left; 0.2s – right). Female and male mice are shown separately. The combined data showed that this modest difference between genotypes in acute seizure threshold was statistically significant (Student's |t|-test, p<0.001). (C) Average latency (# of daily tests) to the first kindled partial seizure following sequential stimulation in B6-+/+ and B6-Ftfl/+ male mice (*Student's |t|-test, p<0.0001). (D) Graph showing the age of onset of observed (behavioral tonic-clonic) seizures in fitful heterozygous mice in the B6 or FVB background. (E) Purkinje cell defect in fitful cerebellum. The upper panels show calbindin antibody staining of wildtype and fitful P15 cerebellum. Notice that the Purkinje cell dendrites are stunted (arrow) in the mutant as compared to wildtype and the soma are less ordered. The lower panels show B6.FVB fitful homozygous and heterozygous Purkinje cells expressing GFP at P17. (F) Auditory brainstem response in fitful and wildtype mice. Left panel: ABR audiograms with average thresholds ± SEM of fitful mouse mutants (open symbols, −/−, n = 14) and their wildtype (n = 5; +/+) and heterozygote (n = 9; +/−) littermates (closed symbols). Right panel: average ABR waveforms (±SEM) of fitful (light grey, n = 9) and wildtype/heterozygous mice (dark grey, n = 9) in response to click stimuli (86 dB, peak equivalent). Latin numbers denominate the ABR peaks.
Figure 2. Conserved protein sequence alignment of Dynamins.
(A) Conserved protein sequence alignment. The upper diagram shows DNM1 sequence conservation among various species and the location of the fitful mutation within the middle domain of the protein. The first splice region (amino acids 399–444) shows the location of the fitful mutation (*408) and the conservation within mammalian, fly and worm dynamins. Notice that the fly and worm orthologs do not have splice variants in this region. The second alternative splicing region is also shown at the end of the PRD domain. GTPase, GTPase domain; Middle, middle domain; PH, pleckstrin homology domain; GED, GTPase effector domain; PRD, proline rich domain. The bottom diagram depicts the mutually exclusive alternative splicing of the Dnm1a and Dnm1b isoforms. (B) Putative orthologues of mouse Dnm1, Dnm2, Dnm3 and Drp (dynamin related protein – official mouse gene symbol, Dnm1l) from different species. For simplicity, the same generic gene symbol is used for all; for simpler eukaryotes, the symbol Dnm refers to their orthologue(s) most like the mammalian Dnm1, Dnm2 or Dnm3. The known or predicted alternate sequences corresponding to mouse Dnm1 exon 10 are also shown. The underlined amino acid symbols show where mouse Dnm1b differs from Dnm1a. The arrow at the bottom shows the highly conserved alanine residue that is mutated to threonine in the mouse Dnm1Ftfl allele. Residues colored in red show amino acid substitutions with respect to mouse Dnm1; for nematode (C.b. - C. briggsae; C.e. – c.elegans), differences between the two Dnm peptides are shown in blue. Peptide sequences were obtained as follows: mouse Dnm1a: GenBank AAA37318, Dnm1b : GenBank EDL08539; human Dnm1a: GenBank AAA02804, Dnm1b: GenBank AAA02803; nematode GenBank AAB72228; fruit fly (D. melanogaster) EMBL CAA42068. The remaining predicted sequences were obtained from analysis using the USCS Genome Browser and draft genome sequence assemblies from the following respective genome centers: chicken (G. gallus), sea lamprey (P. marinus) and nematode (C. briggsae) - Genome Sequencing Center, Washington University School of Medicine; opposum (M. domestica) and lizard (A. carolinensis), The Broad Institute; xenopus (X. tropicalis), lancelet (B. floridae), and sea squirt (C. intestinalis) - DoE Joint Genome Institute; sea urchin (S. purpuratus) - Baylor College of Medicine Human Genome Sequencing Center. The dynamin gene composition from sea lamprey was inferred from sequence alignment of draft 5.9-fold genome sequence (accessed via the UCSC browser - genome.ucsc.edu). When used as query, the respective mouse Dnm1, Dnm2 and Dnm3 peptide sequences corresponding to the assembly domain region each yielded significant alignments with only a single, approximately 20kb contig (Contig16000). This contig contained seven Dnm-like exons, with appropriate splice site recognition motifs, that was co-linear with mouse Dnm1 exons 8–13, including exons corresponding to Dnm1b (exon 10b) and Dnm1a (exon 10a) in the expected 5′-3′ arrangement. (C) Neighbor-joining best tree of dynamin peptides. Proportional number of differences is estimated at the bottom. Note the closer relationship between the two isoforms from sea lamprey and the respective Dnm1a and Dnm1b branches from more complex vertebrates. Also note the closer relationship between Dnm2 isoforms and invertebrate dynamin.
Figure 3. Fitful and compound heterozygous Dnm1 seizure and locomotor phenotypes.
Shown is the frequency of fitful homozygous (Dnm1Ftfl/Dnm1Ftfl) and compound heterozygous (Dnm1Ftfl/Dnm1tm1Pdc) mutant mice and their respective seizure incidence and locomotor phenotypes. The fitful homozygotes shown were from fully informative matings from the mapping cross used to map the recessive phenotype; the compound heterozygotes were from crosses between single heterozygotes from respective FVB-fitful and B6-null colonies. The asterisk is used to indicate that the latter population was observed almost daily from P12 through demise (P16–P23), whereas the former were observed at weaning age only – suggesting that death occurred in these mutants between P12 and P21.
Figure 4. Developmental expression patterns of dynamin-1 isoforms.
(A) mRNA expression levels of Dnm1 isoforms during development in wildtype and mutant animals. The variant isoform region is amplified with common primers and the two transcripts are distinguished by a diagnostic _Hph_I restriction enzyme site specific for the b isoform. The two bands representing the Dnm1b transcript cDNA run as one band and lower on the gel than the Dnm1a transcript cDNA. Note that the 1a isoform becomes increasingly upregulated during development, while the b isoform is down regulated, however this ratio of a to b transcripts is skewed in the mutant animals. Below the gel is a schematic outlining the basis of the assay. Additional replicates are shown in Figure S2. (B) Left, quantification of Dnm1b mRNA expression as assayed by PCR and visualized on agarose gels, relative to total Dnm1 mRNA in whole brain of E17.5, P0, and P14 wildtype and mutant brains. Data are expressed as the mean (± SEM) relative proportion of Dnm1b to Dnm1 mRNA in the total cleaved PCR product. *P<0.05, Student's t-test. Right, relative quantification of isoform transcripts by pyrosequencing. cDNA from P14 wildtype or homozygous fitful brains was amplified by PCR, and subsequently analyzed by pyrosequencing and quantitated for Dnm1b levels relative to Dnm1; n = 3 for each set of cDNA. (C) Protein levels of Dnm1 isoforms during development in wildtype and mutant animals. Custom made antibodies that distinguish DNM1 exon10a and exon10b middle domains were used to assay isoform protein levels in brain extracts of mice during development. Note the decrease in DNM1A in homozygous fitful mice compared to wildtype at all three ages examined (P7, P11, P15). Overall DNM1 protein levels were detected with a commercial antibody against full length DNM1. For a loading control, β-tubulin levels were detected. Significant difference in levels of DNM1b in mutant (Least square regression analysis; P<0.05).
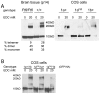
Figure 5. DNM1aFtfl is defective in higher order homo-oligomerization.
(A) Left panel, protein extract from P14 whole brain tissue of homozygous fitful and wildtype littermates incubated with 0 or 20mM EDC cross-linker and hybridized with anti-dynamin-1 antibody. Monomers migrate at 100kD, dimers at 200kD and the tetramers are at 400kD. This assay was performed over three separate times with different samples each time; a representative blot with corresponding percentages is shown. Mean densities (± 1SD) from all experiments are: wildtype 28.75±8.24 (monomer), 29.67±13.9 (dimer), 43.9±8.5 (tetramer); mutant 44±10.7 (monomer), 39.5±12.2 (dimer), 23±13.5 (tetramer) Right panel, COS-7 cells transfected with DNM1-GFP constructs show differences in dimerization. (B) COS-7 cells doubly transfected with DNM1-GFP and DNM1-HA constructs show isoform heterodimerization. Protein extracts from cells were incubated with 0 or 20mM EDC and analyzed by Western blot. Blots were hybridized with anti-GFP antibody, stripped of antibody and then re-hybridized with anti-HA antibody in order to ascertain the presence of each construct in the dimers. A representative blot hybridized with anti-GFP antibody is shown.
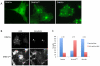
Figure 6. DNM1 localization and endocytosis in COS-7 cells.
(A) DNM1 isoform localization in COS-7 cells over-expressing DNM1 isoform constructs. Left is wildtype, middle is DNM1aFtfl and right is DNM1b. Note the tubulation in the mutant expressing cells. (B) Transferrin endocytosis and localization in COS-7 cells overexpressing DNM1 isoform constructs. Left panels show DNM1 GFP fluorescence in COS-7 cells containing wildtype or DNM1aFtfl; right panels show TRITC-transferrin uptake and localization in COS-7 cells containing wildtype or DNM1aFtfl -GFP. The arrowheads in the top right picture indicate the normal perinuclear accumulation of transferrin. The arrows in the bottom right picture indicate one point of co-localization between DNM1aFtfl and transferrin. (C) Quantification of transferrin localization in COS-7 cells overexpressing DNM1 isoform constructs. Transfected cells were determined to have transferrin localized to the region adjacent to the nucleus in a manner similar to non-transfected cells or to have not taken up transferrin at all or to have an abnormal localization of transferrin.
Figure 7. Altered GABAergic transmission in cortical neurons of fitful mice.
(A) Samples of GABAergic mIPSCs recorded from a wildtype (upper trace) and a fitful (lower trace) layer V pyramidal neuron at P14 in the presence of DNQX, kynurenic acid, and TTX. The patch pipettes contained 130 mM KCl, and holding potential was at −70 mV. (B) Averaged mIPSCs from 21 wildtype cells (grey) and 22 fitful cells (red) with normalized peaks. (C,D) Histograms of the decay constant (C) and rise time (D) of mIPSCs for wildtype and fitful cells. The mean decay time constant was 5.2±0.2 ms (n = 22) for mutant cells, and 4.1±0.2 ms (n = 21) for wildtype (p<0.005, Mann-Whitney test). (E) Samples of evoked GABAergic IPSCs in response to 10-Hz stimulation recorded from a wildtype (upper black traces) and a fitful (lower red traces) cell at P14 in the presence of DNQX and kynurenic acid. For both cells, the three traces were responses to the first, 100th, and 500th stimulus. The patch pipettes contained 110 mM cesium methylsulfate, and holding potential was at 0 mV. A pair of twisted microwire was placed in layer V to evoke monosynaptic IPSCs. (F) Plots of evoked IPSCs in response to 400 stimuli at 10 Hz. For each cell, peak amplitudes of IPSCs were normalized to that of the baseline response. (G) Recovery after 1000 stimuli at 10 Hz. IPSCs were measured every 10 s after the 10-Hz stimulation. Peak amplitudes of IPSCs were normalized to that of the baseline response before the 10-Hz train stimulation.
Similar articles
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy.
Asinof SK, Sukoff Rizzo SJ, Buckley AR, Beyer BJ, Letts VA, Frankel WN, Boumil RM. Asinof SK, et al. PLoS Genet. 2015 Jun 30;11(6):e1005347. doi: 10.1371/journal.pgen.1005347. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26125563 Free PMC article. - Dynamin 1 isoform roles in a mouse model of severe childhood epileptic encephalopathy.
Asinof S, Mahaffey C, Beyer B, Frankel WN, Boumil R. Asinof S, et al. Neurobiol Dis. 2016 Nov;95:1-11. doi: 10.1016/j.nbd.2016.06.014. Epub 2016 Jun 28. Neurobiol Dis. 2016. PMID: 27363778 Free PMC article. - A deep intronic variant in DNM1 in a patient with developmental and epileptic encephalopathy creates a splice acceptor site and affects only transcript variants including exon 10a.
Harms FL, Weiss D, Lisfeld J, Alawi M, Kutsche K. Harms FL, et al. Neurogenetics. 2023 Jul;24(3):171-180. doi: 10.1007/s10048-023-00716-w. Epub 2023 Apr 11. Neurogenetics. 2023. PMID: 37039969 - Abnormal axonal development and severe epileptic phenotype in Dynamin-1 (DNM1) encephalopathy.
Matsubara K, Kuki I, Ishioka R, Yamada N, Fukuoka M, Inoue T, Nukui M, Okamoto N, Mizuguchi T, Matsumoto N, Okazaki S. Matsubara K, et al. Epileptic Disord. 2024 Feb;26(1):139-143. doi: 10.1002/epd2.20181. Epub 2023 Dec 2. Epileptic Disord. 2024. PMID: 38009673 Review. - [Dynamin-1-related infantile spasms: a case report and review of literature].
Deng XL, Yin F, Zhang CL, Ma YP, He F, Wu LW, Peng J. Deng XL, et al. Zhonghua Er Ke Za Zhi. 2016 Nov 2;54(11):856-859. doi: 10.3760/cma.j.issn.0578-1310.2016.11.014. Zhonghua Er Ke Za Zhi. 2016. PMID: 27806796 Review. Chinese.
Cited by
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy.
Asinof SK, Sukoff Rizzo SJ, Buckley AR, Beyer BJ, Letts VA, Frankel WN, Boumil RM. Asinof SK, et al. PLoS Genet. 2015 Jun 30;11(6):e1005347. doi: 10.1371/journal.pgen.1005347. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26125563 Free PMC article. - Proper synaptic vesicle formation and neuronal network activity critically rely on syndapin I.
Koch D, Spiwoks-Becker I, Sabanov V, Sinning A, Dugladze T, Stellmacher A, Ahuja R, Grimm J, Schüler S, Müller A, Angenstein F, Ahmed T, Diesler A, Moser M, Tom Dieck S, Spessert R, Boeckers TM, Fässler R, Hübner CA, Balschun D, Gloveli T, Kessels MM, Qualmann B. Koch D, et al. EMBO J. 2011 Sep 16;30(24):4955-69. doi: 10.1038/emboj.2011.339. EMBO J. 2011. PMID: 21926968 Free PMC article. - Association between the polymorphisms in intercellular adhesion molecule-1 and the risk of coronary atherosclerosis: a case-controlled study.
Yang M, Fu Z, Zhang Q, Xin Y, Chen Y, Tian Y. Yang M, et al. PLoS One. 2014 Oct 13;9(10):e109658. doi: 10.1371/journal.pone.0109658. eCollection 2014. PLoS One. 2014. PMID: 25310099 Free PMC article. - The DyNaMics of Excitation and Inhibition Govern Epileptic Encephalopathies and Their Comorbidities.
Weston M. Weston M. Epilepsy Curr. 2016 May-Jun;16(3):172-3. doi: 10.5698/1535-7511-16.3.172. Epilepsy Curr. 2016. PMID: 27330446 Free PMC article. No abstract available. - Let's go bananas: revisiting the endocytic BAR code.
Qualmann B, Koch D, Kessels MM. Qualmann B, et al. EMBO J. 2011 Aug 31;30(17):3501-15. doi: 10.1038/emboj.2011.266. EMBO J. 2011. PMID: 21878992 Free PMC article. Review.
References
- Berkovic SF, Mulley JC, Scheffer IE, Petrou S. Human epilepsies: interaction of genetic and acquired factors. Trends Neurosci. 2006;29:391–397. - PubMed
- Gardiner M. Genetics of idiopathic generalized epilepsies. Epilepsia. 2005;46(Suppl 9):15–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 NS031348/NS/NINDS NIH HHS/United States
- NS31348/NS/NINDS NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- R01 NS031348/NS/NINDS NIH HHS/United States
- R01 NS064013/NS/NINDS NIH HHS/United States
- R03 NS065255/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous