Single-molecule derivation of salt dependent base-pair free energies in DNA - PubMed (original) (raw)
Single-molecule derivation of salt dependent base-pair free energies in DNA
Josep M Huguet et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Accurate knowledge of the thermodynamic properties of nucleic acids is crucial to predicting their structure and stability. To date most measurements of base-pair free energies in DNA are obtained in thermal denaturation experiments, which depend on several assumptions. Here we report measurements of the DNA base-pair free energies based on a simplified system, the mechanical unzipping of single DNA molecules. By combining experimental data with a physical model and an optimization algorithm for analysis, we measure the 10 unique nearest-neighbor base-pair free energies with 0.1 kcal mol(-1) precision over two orders of magnitude of monovalent salt concentration. We find an improved set of standard energy values compared with Unified Oligonucleotide energies and a unique set of 10 base-pair-specific salt-correction values. The latter are found to be strongest for AA/TT and weakest for CC/GG. Our unique energy values and salt corrections improve predictions of DNA unzipping forces and are fully compatible with melting temperatures for oligos. The method should make it possible to obtain free energies, enthalpies, and entropies in conditions not accessible by bulk methodologies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
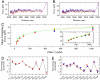
Experimental setup and results. (A) Molecular construct. A sequence of 6,838 bp obtained from _λ_-DNA is ligated to a tetraloop and two short handles of 29 bp each. (B) Experimental setup. The molecular construct is attached to two beads. The unzipping experiment is performed by moving the optical trap relative to the pipette. (C) Unfolfing (red) and refolding (green) curves filtered with a running average filter of 1 Hz bandwidth. There is always some hysteresis in the last rip. Pulling and relaxing are almost identical in the rest of the curve. Inset shows raw data (same color as before, blue curve is the average at bandwidth 1 Hz). (D) FDCs at various monovalent salt concentrations. (E) Elastic response of a 3 kb ssDNA molecule at various salts. Raw data of three molecules are shown (orange, green and blue curves). Red curve shows the best-fit to the elastic model.
Fig. 2.
Salt dependencies. (A, B) FDCs for the 6.8 kb sequence at 10 mM NaCl (A) and 1 M NaCl (B). Black curve, experimental measurements; blue curve, UO prediction; red curve, our fit; magenta curve, elastic response of the fully unzipped molecule. The theoretical FDC is calculated in equilibrium, which assumes that the bandwidth is 0 Hz and the experimental data is filtered at bandwidth 1 Hz. If data is filtered at higher frequencies (> 1 Hz), hopping between states is observed and the experimental FDC does not compare well with the theoretical FDC at equilibrium. If data is filtered at lower frequencies (< 1 Hz), the force rips are smoothed and hopping transitions are averaged out. (C) Dependence of mean unzipping force with salt concentration. Red points, experimental measurements for the 6.8 kb sequence; green curve, UO prediction for the 6.8 kb sequence; blue points, experimental measurements for the 2.2 kb sequence; orange curve, UO prediction for the 2.2 kb sequence. (D, E) NNBP energies and comparison with UO values at 10 mM NaCl (D) and 1 M NaCl (E). The following notation is used for NNBP: AG/TC denotes 5′-AG-3′ paired with 5′-CT-3′. Black points, UO values; red points, values for the 6.8 kb molecule; blue points, values for the 2.2 kb molecule. The values for the 6.8 kb and the 2.2 kb molecules have been obtained after averaging over six molecules. Error bars are determined from the standard error among different molecules.
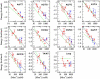
Fig. 3.
Salt corrections of the NNBP energies. Figure shows the energy of all different NNBP parameters. Red (blue) points are the experimental results for the 6.8 kb (2.2 kb) sequence; green curve, UO nonspecific salt correction; black curve, fit to Eq. 3 with adjustable parameters m i (i = 1,…,10, loop) and  .
.
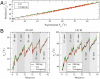
Fig. 4.
Melting temperatures prediction. Comparison with melting temperatures for the 92 oligos ranging from 10–30 bp reported in ref. (data reported in
SI Appendix: Table S2
). (A) Predicted vs. experimentally measured melting temperatures at five salt conditions ([Na+] = 69, 119, 220, 621, and 1,020 mM). The values obtained from unzipping have less error at higher temperatures (corresponding to longer oligos). (B) Prediction at 69 mM (left) and 1.02 M NaCl (right). Black lines are the experimentally measured melting temperatures, green line is the UO prediction and red line our prediction from unzipping data.
Similar articles
- Derivation of nearest-neighbor DNA parameters in magnesium from single molecule experiments.
Huguet JM, Ribezzi-Crivellari M, Bizarro CV, Ritort F. Huguet JM, et al. Nucleic Acids Res. 2017 Dec 15;45(22):12921-12931. doi: 10.1093/nar/gkx1161. Nucleic Acids Res. 2017. PMID: 29177444 Free PMC article. - Heat capacity effects on the melting of DNA. 2. Analysis of nearest-neighbor base pair effects.
Rouzina I, Bloomfield VA. Rouzina I, et al. Biophys J. 1999 Dec;77(6):3252-5. doi: 10.1016/S0006-3495(99)77156-0. Biophys J. 1999. PMID: 10585947 Free PMC article. - Stability of DNA duplexes containing GG, CC, AA, and TT mismatches.
Tikhomirova A, Beletskaya IV, Chalikian TV. Tikhomirova A, et al. Biochemistry. 2006 Sep 5;45(35):10563-71. doi: 10.1021/bi060304j. Biochemistry. 2006. PMID: 16939208 - Nucleic Acid Thermodynamics Derived from Mechanical Unzipping Experiments.
Rissone P, Ritort F. Rissone P, et al. Life (Basel). 2022 Jul 20;12(7):1089. doi: 10.3390/life12071089. Life (Basel). 2022. PMID: 35888177 Free PMC article. Review. - Non-linear Hamiltonian models for DNA.
Zoli M. Zoli M. Eur Biophys J. 2022 Sep;51(6):431-447. doi: 10.1007/s00249-022-01614-z. Epub 2022 Aug 17. Eur Biophys J. 2022. PMID: 35976412 Review.
Cited by
- Recent developments in single-molecule DNA mechanics.
Bryant Z, Oberstrass FC, Basu A. Bryant Z, et al. Curr Opin Struct Biol. 2012 Jun;22(3):304-12. doi: 10.1016/j.sbi.2012.04.007. Epub 2012 May 31. Curr Opin Struct Biol. 2012. PMID: 22658779 Free PMC article. Review. - Improving signal/noise resolution in single-molecule experiments using molecular constructs with short handles.
Forns N, de Lorenzo S, Manosas M, Hayashi K, Huguet JM, Ritort F. Forns N, et al. Biophys J. 2011 Apr 6;100(7):1765-74. doi: 10.1016/j.bpj.2011.01.071. Biophys J. 2011. PMID: 21463590 Free PMC article. - The shaping of a molecular linguist: How a career studying DNA energetics revealed the language of molecular communication.
Breslauer KJ. Breslauer KJ. J Biol Chem. 2021 Jan-Jun;296:100522. doi: 10.1016/j.jbc.2021.100522. Epub 2021 Apr 7. J Biol Chem. 2021. PMID: 34237886 Free PMC article. - Predictability of environment-dependent formation of G-quadruplex DNAs in human mitochondria.
Liu L, Takahashi S, Ghosh S, Endoh T, Yoshinaga N, Numata K, Sugimoto N. Liu L, et al. Commun Chem. 2025 May 3;8(1):135. doi: 10.1038/s42004-025-01532-z. Commun Chem. 2025. PMID: 40319099 Free PMC article. - Stabilization of DNA fork junctions by Smc5/6 complexes revealed by single-molecule imaging.
Tanasie NL, Gutiérrez-Escribano P, Jaklin S, Aragon L, Stigler J. Tanasie NL, et al. Cell Rep. 2022 Dec 6;41(10):111778. doi: 10.1016/j.celrep.2022.111778. Cell Rep. 2022. PMID: 36476856 Free PMC article.
References
- Calladine C, Drew H, Luisi B, Travers A. Understanding DNA: the molecule and how it works. 3rd Ed. Amsterdam: Elsevier Academic Press; 2004.
- Devoe H, Tinoco IJ. The stability of helical polynucleotides: base contributions. J Mol Biol. 1962;4:500–517. - PubMed
- Crothers DM, Zimm BH. Theory of the melting transition of synthetic polynucleotides: evaluation of the stacking free energy. J Mol Biol. 1964;9:1–9. - PubMed
- Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources