Comprehensive methylome map of lineage commitment from haematopoietic progenitors - PubMed (original) (raw)
. 2010 Sep 16;467(7313):338-42.
doi: 10.1038/nature09367. Epub 2010 Aug 15.
Lauren I R Ehrlich, Jun Seita, Peter Murakami, Akiko Doi, Paul Lindau, Hwajin Lee, Martin J Aryee, Rafael A Irizarry, Kitai Kim, Derrick J Rossi, Matthew A Inlay, Thomas Serwold, Holger Karsunky, Lena Ho, George Q Daley, Irving L Weissman, Andrew P Feinberg
Affiliations
- PMID: 20720541
- PMCID: PMC2956609
- DOI: 10.1038/nature09367
Comprehensive methylome map of lineage commitment from haematopoietic progenitors
Hong Ji et al. Nature. 2010.
Abstract
Epigenetic modifications must underlie lineage-specific differentiation as terminally differentiated cells express tissue-specific genes, but their DNA sequence is unchanged. Haematopoiesis provides a well-defined model to study epigenetic modifications during cell-fate decisions, as multipotent progenitors (MPPs) differentiate into progressively restricted myeloid or lymphoid progenitors. Although DNA methylation is critical for myeloid versus lymphoid differentiation, as demonstrated by the myeloerythroid bias in Dnmt1 hypomorphs, a comprehensive DNA methylation map of haematopoietic progenitors, or of any multipotent/oligopotent lineage, does not exist. Here we examined 4.6 million CpG sites throughout the genome for MPPs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), and thymocyte progenitors (DN1, DN2, DN3). Marked epigenetic plasticity accompanied both lymphoid and myeloid restriction. Myeloid commitment involved less global DNA methylation than lymphoid commitment, supported functionally by myeloid skewing of progenitors following treatment with a DNA methyltransferase inhibitor. Differential DNA methylation correlated with gene expression more strongly at CpG island shores than CpG islands. Many examples of genes and pathways not previously known to be involved in choice between lymphoid/myeloid differentiation have been identified, such as Arl4c and Jdp2. Several transcription factors, including Meis1, were methylated and silenced during differentiation, indicating a role in maintaining an undifferentiated state. Additionally, epigenetic modification of modifiers of the epigenome seems to be important in haematopoietic differentiation. Our results directly demonstrate that modulation of DNA methylation occurs during lineage-specific differentiation and defines a comprehensive map of the methylation and transcriptional changes that accompany myeloid versus lymphoid fate decisions.
Figures
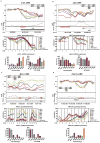
Figure 1. Examples of known lineage-related genes showing differential DNA methylation between lymphoid and myeloid progenitors
a, Hematopoietic progenitors included in this study. Dashed-arrow indicates existence of intermediate progenitors. DMR in b, Lck and c, Mpo. Upper panels: top half: CpG methylation (p); lower half: CpG dinucleotides (black tick marks), CpG density (curve), CpG islands (orange lines) and the gene annotation (see online Methods). Middle panels: methylation of individual CpGs (in the red boxes), mean values connected by lines. Bottom panels: mRNA expression levels, normalized to the highest expression among the populations (mean ± s.d., n=3; 5 for MPPFL− for microarrays).
Figure 2. Gene expression correlates strongly with DMRs at shores
DMRs within 2kb of gene TSSs (black circles) were divided into two groups: Island (inside, cover, or overlap more than 50% of a CpG island), and Shores (up to 2000bp away from a CpG island). After RMA preprocessing, the log2 ratios of the gene expression differences (from leftto right) were plotted against Δp (left group minus right group). Black pluses represent random DMR-gene pairs more than 2kb apart. Wilcoxon rank-sum tests were performed to test the null hypothesis. a, MPPFL− vs. DN3_DMRs. b, MPPFL− vs. GMP_DMRs.
Figure 3. CHARM identified genes with previously unknown functions in lymphoid/myeloid lineage commitment and pluripotency maintenance
a and b, Examples of DMRs with methylation changes in lymphoid/myeloid progenitors. a, the DMR in Arl4c. b, the DMR in Jdp2. C, the DMR in Meis1. D, the DMR in Hdac7a. The CHARM plots, pyrosequencing, Affymetrix GeneChip, and RT-PCR data are organized and displayed as in Fig. 1b.
Comment in
- Stem cells: Troublesome memories.
Zwaka TP. Zwaka TP. Nature. 2010 Sep 16;467(7313):280-1. doi: 10.1038/467280a. Nature. 2010. PMID: 20844526 No abstract available.
Similar articles
- Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment.
Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Miyamoto T, et al. Dev Cell. 2002 Jul;3(1):137-47. doi: 10.1016/s1534-5807(02)00201-0. Dev Cell. 2002. PMID: 12110174 - DNA Methylation Dynamics of Human Hematopoietic Stem Cell Differentiation.
Farlik M, Halbritter F, Müller F, Choudry FA, Ebert P, Klughammer J, Farrow S, Santoro A, Ciaurro V, Mathur A, Uppal R, Stunnenberg HG, Ouwehand WH, Laurenti E, Lengauer T, Frontini M, Bock C. Farlik M, et al. Cell Stem Cell. 2016 Dec 1;19(6):808-822. doi: 10.1016/j.stem.2016.10.019. Epub 2016 Nov 17. Cell Stem Cell. 2016. PMID: 27867036 Free PMC article. - IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors.
Becker AM, Michael DG, Satpathy AT, Sciammas R, Singh H, Bhattacharya D. Becker AM, et al. Blood. 2012 Mar 1;119(9):2003-12. doi: 10.1182/blood-2011-06-364976. Epub 2012 Jan 11. Blood. 2012. PMID: 22238324 Free PMC article. - Biological and molecular evidence for existence of lymphoid-primed multipotent progenitors.
Luc S, Buza-Vidas N, Jacobsen SE. Luc S, et al. Ann N Y Acad Sci. 2007 Jun;1106:89-94. doi: 10.1196/annals.1392.023. Epub 2007 Apr 18. Ann N Y Acad Sci. 2007. PMID: 17442777 Review. - Role of transcription factors in differentiation and reprogramming of hematopoietic cells.
Nakajima H. Nakajima H. Keio J Med. 2011;60(2):47-55. doi: 10.2302/kjm.60.47. Keio J Med. 2011. PMID: 21720200 Review.
Cited by
- Dynamic DNA methylation across diverse human cell lines and tissues.
Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Varley KE, et al. Genome Res. 2013 Mar;23(3):555-67. doi: 10.1101/gr.147942.112. Epub 2013 Jan 16. Genome Res. 2013. PMID: 23325432 Free PMC article. - Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia.
Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, Kweon SM, Zickl L, Shojaee S, Neuberg D, Huang C, Biswas D, Xin Y, Racevskis J, Ketterling RP, Luger SM, Lazarus H, Tallman MS, Rowe JM, Litzow MR, Guzman ML, Allis CD, Roeder RG, Müschen M, Paietta E, Elemento O, Melnick AM. Geng H, et al. Cancer Discov. 2012 Nov;2(11):1004-23. doi: 10.1158/2159-8290.CD-12-0208. Epub 2012 Oct 29. Cancer Discov. 2012. PMID: 23107779 Free PMC article. - Genomic insights into cancer-associated aberrant CpG island hypermethylation.
Sproul D, Meehan RR. Sproul D, et al. Brief Funct Genomics. 2013 May;12(3):174-90. doi: 10.1093/bfgp/els063. Epub 2013 Jan 21. Brief Funct Genomics. 2013. PMID: 23341493 Free PMC article. Review. - Plasticity of DNA methylation in mouse T cell activation and differentiation.
Li Y, Chen G, Ma L, Ohms SJ, Sun C, Shannon MF, Fan JY. Li Y, et al. BMC Mol Biol. 2012 May 29;13:16. doi: 10.1186/1471-2199-13-16. BMC Mol Biol. 2012. PMID: 22642378 Free PMC article. - Dysregulation of epigenetic modifications in inborn errors of immunity.
Xiao Z, He R, Zhao Z, Chen T, Ying Z. Xiao Z, et al. Epigenomics. 2024;16(19-20):1301-1313. doi: 10.1080/17501911.2024.2410695. Epub 2024 Oct 15. Epigenomics. 2024. PMID: 39404224 Review.
References
- Broske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. - PubMed
- Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. - PubMed
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. - PubMed
Publication types
MeSH terms
Grants and funding
- R01AI047458/AI/NIAID NIH HHS/United States
- T32 AI007290/AI/NIAID NIH HHS/United States
- P50 HG003233/HG/NHGRI NIH HHS/United States
- R01 GM083084-04/GM/NIGMS NIH HHS/United States
- R01 AI047457-05/AI/NIAID NIH HHS/United States
- T32 CA009151/CA/NCI NIH HHS/United States
- R37 CA054358-18/CA/NCI NIH HHS/United States
- R00 AG029760/AG/NIA NIH HHS/United States
- R37CA053458/CA/NCI NIH HHS/United States
- F32AI058521/AI/NIAID NIH HHS/United States
- CA09151/CA/NCI NIH HHS/United States
- R01 AI047457-04/AI/NIAID NIH HHS/United States
- F32 AI058521-02/AI/NIAID NIH HHS/United States
- R01 CA086065/CA/NCI NIH HHS/United States
- P50 HG003233-07/HG/NHGRI NIH HHS/United States
- R37 CA054358/CA/NCI NIH HHS/United States
- P50 HG003233-08/HG/NHGRI NIH HHS/United States
- R01 GM083084/GM/NIGMS NIH HHS/United States
- R01 AI047458/AI/NIAID NIH HHS/United States
- R01AI047457/AI/NIAID NIH HHS/United States
- R00 AG029760-04/AG/NIA NIH HHS/United States
- R37 CA054358-19/CA/NCI NIH HHS/United States
- P50HG003233/HG/NHGRI NIH HHS/United States
- F32 AI058521/AI/NIAID NIH HHS/United States
- R01 AI047457/AI/NIAID NIH HHS/United States
- R00AGO29760/PHS HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous